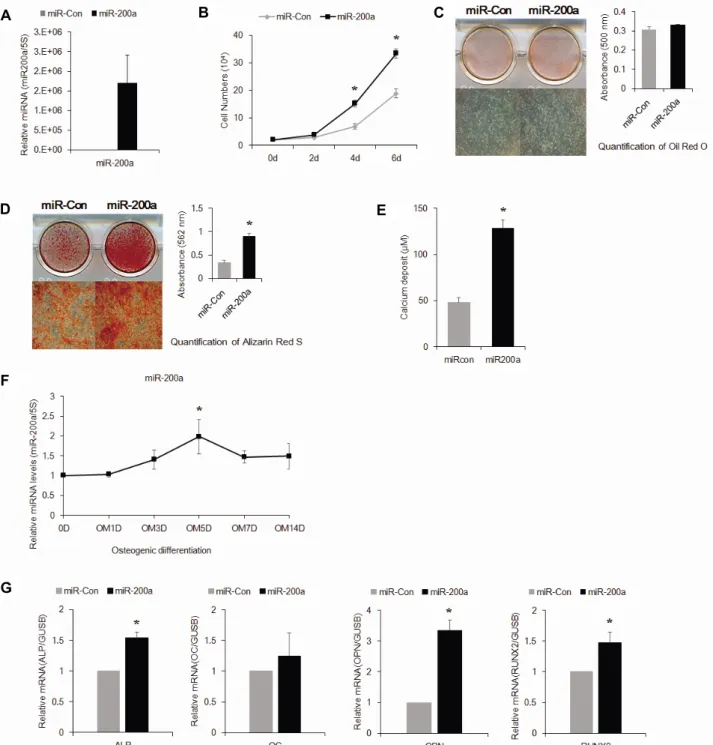
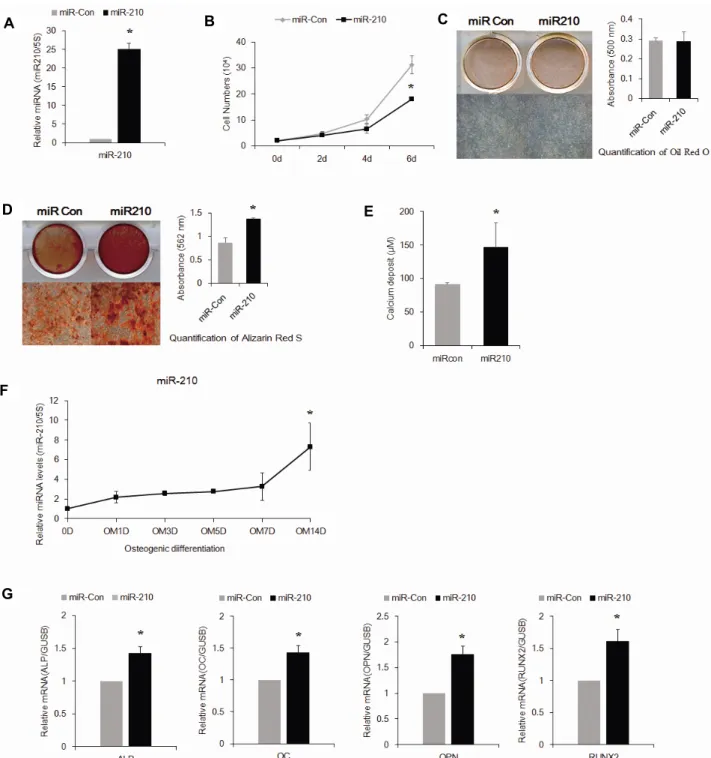
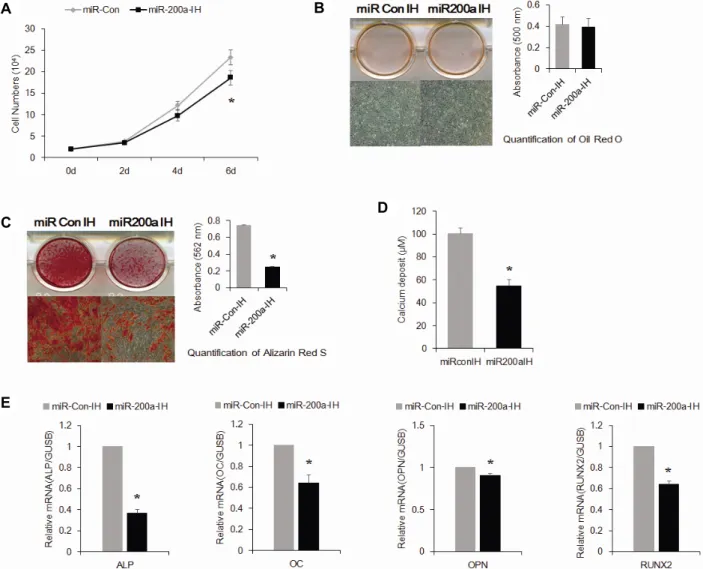
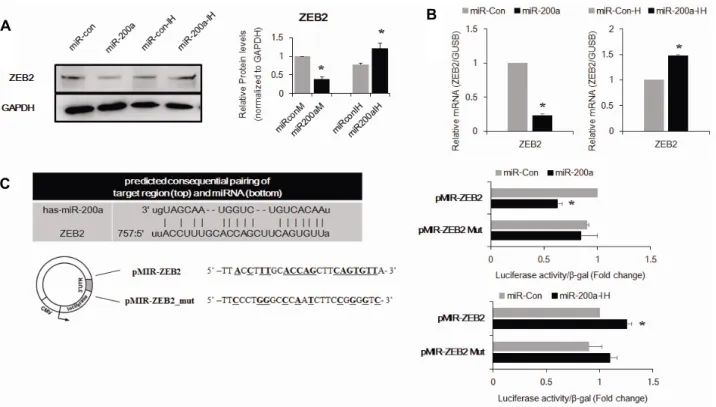
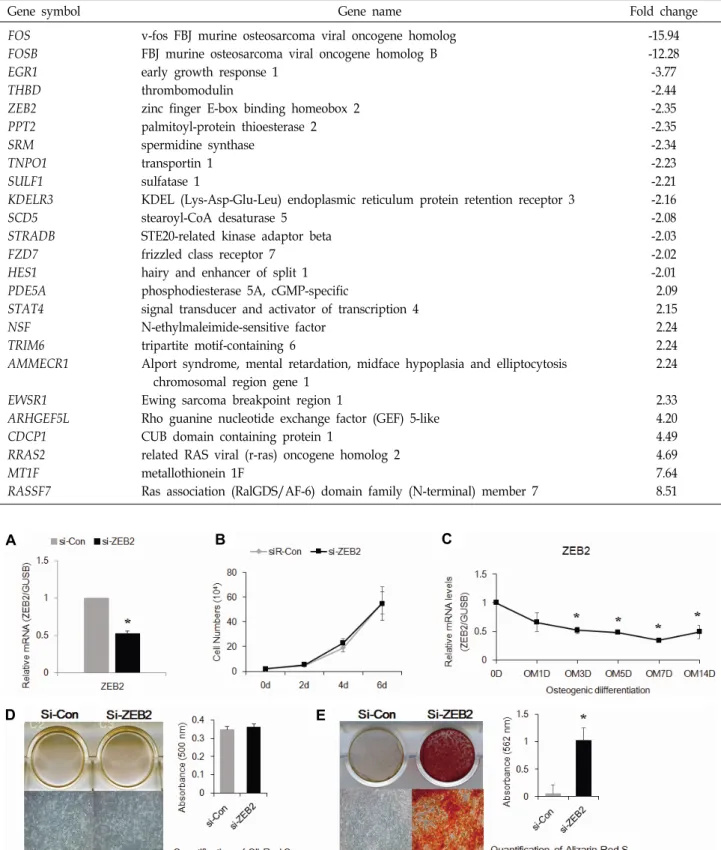
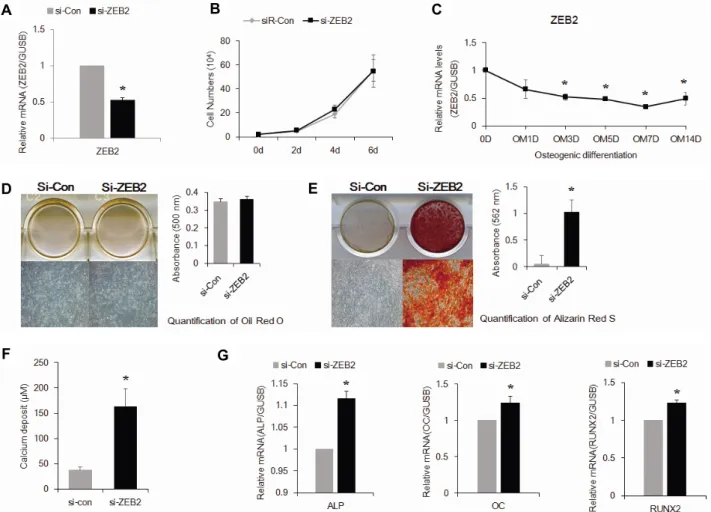
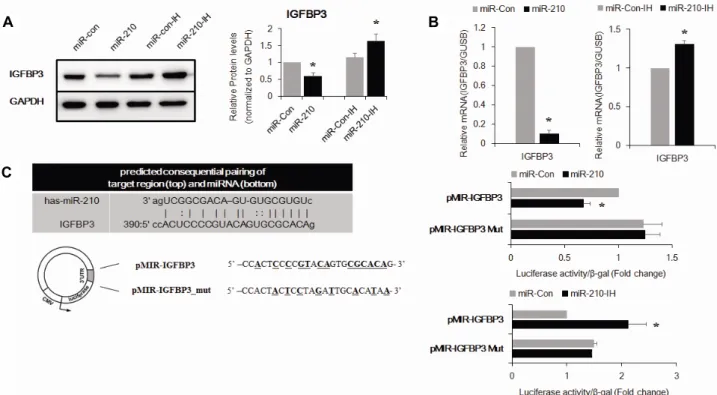
관련 문서
The surface of untreated Ti, SLA treated Ti, and AA plasma polymerized SLA/Ti were also examined for their proliferation and differentiation of
In this study, we have fabricated the 660 nm red LED illumination controller and LED array module and evaluated the proliferation and differentiation
Determining optimal surface roughness of TiO₂blasted titanium implant material for attachment, proliferation and differentiation of cells derived from
Conclusions: Thus, 25-HC induced odontoclast differentiation through the odontoclastogenesis factor-mediated activation of NF-κB and upregulated inflammatory
3 An Analytic Function of Constant Absolute Value Is Constant The Cauchy-Riemann equations also help in deriving general. properties
These results of the histologic distribution of lamina propria, adipose tissue, and glandular tissue of the hard palatal mucosa and the topography of the greater palatine
differentiation, integration (line, surface and volume differentiation, integration (line, surface and volume integrals).. del( ) operator, gradient, divergence and
Fig 4 : Inhibitory effects of classified methanol extracts of Acalypha australis the COX-2 protein expression of the human oral cavity carcinoma KB cells....