Correspondence to: Hyung Hwan Baik, Kohwang Medical Research Institute, Department of Biochemistry and Molecular Biology, School of Medicine, Kyung Hee University, 1, Hoegi-dong, Dongdaemun-gu, Seoul 130-701, Korea
Tel: +82-2-961-0317, E-mail: hhbaik@khu.ac.kr
Received May 15, 2009, Revised May 30, 2009, Accepted June 10, 2009.
SNPs and Haplotypes of Ets variant 6 (ETV6) Gene Is Associated with Overweight/Obese in Korean Population
*Kohwang Medical Research Institute, School of Medicine, Kyung Hee University,
†Kyung Hee University East-West Neo Medical Center, Seoul, Korea
Kyung Hee Jung*, Sung Wook Kang*, Su Kang Kim*, Jeong Yoon Song†, Hyung Kyung Park†, Hyung Hwan Baik*
Ets variant 6 (ETV6) gene is known to be involved in a large number of chromosomal rearrangements associated with leukemia.
In recent study, it is reported that excess body weight may be a risk factor for leukemia and overweight/obesity have greater risk of leukemia compared to non-overweight individuals. For a possible relation of the genetic polymorphism of ETV6 with overweight/obesity, we genotyped fifty-five single nucleotide polymorphisms (SNPs) of ETV6 using the Affymetrix Genotyping chip array in 206 overweight/obese and 152 normal subjects in the Korean population. Logistic and multiple regression models were employed to analyze the genetic contributions of polymorphisms. Of the fifty five polymorphisms, the four SNPs showed a significant association with overweight/obesity in 3 alternative models (codominant, dominant, and recessive models; p<0.05 after adjusting for age and sex) were rs2723825, rs17818048, rs2710304, and rs7296981. Two haplotypes of ETV6 (block 7: GGG;
block 9: CCC) containing significant polymorphisms (rs17818048, rs2710304, and rs7296981) exhibited significant association with the risk of overweight/obesity. Our results suggest that polymorphisms in ETV6 may be associated with overweight/obese in Korean population. (Korean J Str Res 2009;17:163∼168)
Key Words: Ets variant 6 (ETV6), Haplotype, Obesity, Single nucleotide polymorphism (SNP)
INTRODUCTION
Obesity is the most prevalent nutritional disorder that results of imbalance between energy intake and expenditure. It is often associated with hyperlipidemia, type 2 diabetes mellitus, and hypertension and with increased risk of coronary heart disease
(Leonhardt et al., 1999). Emerging epidemiologic evidence also suggests that excess overweight/obese may be a risk factor for hematologic cancers, including non-Hodgkin’s lymphoma, multiple myeloma possibly leukemia (Larsson et al., 2007a). It is revealed that excess body weight may be a risk factor for leukemia and overweight/obese individuals have greater risk of leukemia compared to non-overweight individuals. Obesity was directly associated with the risk of leukemia in both men and women and with all subtypes leukemia (Larsson et al., 2007b).
Ets variant 6 (ETV6) gene on 12p13, known as translocation ets leukemia (TEL) encodes an E-26 transforming specific (ETS) family transcription factor with two functional domains: C-terminal
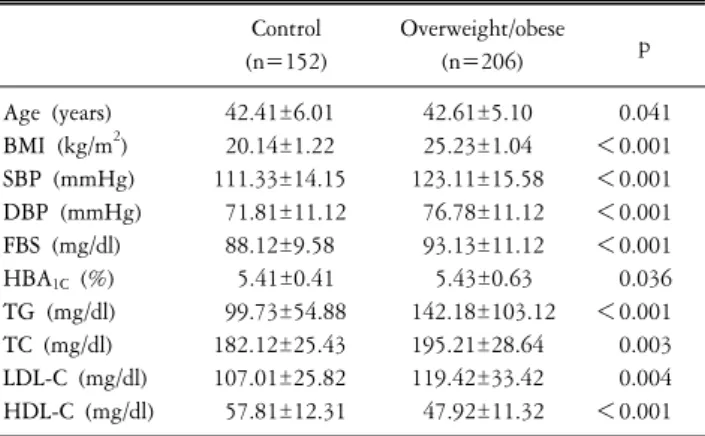
Table 1. Clinical characteristics of overweight/obese and control subjects.
Control Overweight/obese
(n=152) (n=206) p
Age (years) 42.41±6.01 42.61±5.10 0.041
BMI (kg/m2) 20.14±1.22 25.23±1.04 <0.001
SBP (mmHg) 111.33±14.15 123.11±15.58 <0.001
DBP (mmHg) 71.81±11.12 76.78±11.12 <0.001 FBS (mg/dl) 88.12±9.58 93.13±11.12 <0.001 HBA1C (%) 5.41±0.41 5.43±0.63 0.036 TG (mg/dl) 99.73±54.88 142.18±103.12 <0.001
TC (mg/dl) 182.12±25.43 195.21±28.64 0.003
LDL-C (mg/dl) 107.01±25.82 119.42±33.42 0.004 HDL-C (mg/dl) 57.81±12.31 47.92±11.32 <0.001 Data are means±SD. BMI: body mass index, SBP: systolic blood pressure, DBP: diastolic blood pressure, FBS: fasting plasma glucose, HBA1C: hemoglobin A1c, TG: triglyceride, TC: total cholesterol, LDL-C:
low-density lipoprotein, HDL-C: high-density lipoprotein.
DNA-binding domain that recognizes ETS binding sites in promoter regions of target genes (Fenrick et al., 2000) and N-terminal pointed homodimerization domain (Golub et al., 2000). This gene is known to be involved in a large number of chromosomal rearrangements associated with leukemia and congenital fibrosarcoma (Wlodarska et al., 1996; Odero et al., 2001). In recent, there were investigated that ETV6 abnormalities are not restricted to translocations and occur more frequently in acute myeloid leukemia (AML) (Barjesteck et al., 2005). However, there are no studies so far to investigate the association between single nucleotide polymorphisms (SNPs) of the ETV6 gene and overweight/obese in the Korean population. We first searched for the genetic variation in ETV6 gene by downloading all SNPs typed from the HapMap database, and selected 55 SNPs. And we investigated whether such genetic polymorphisms of ETV6 gene are associated with the overweight/obesity as measured by body mass index (BMI) in Korean population.
MATERIALS AND METHODS
1. Subjects and DNA samples
The body mass index (BMI) was calculated from height and weight using the formula: BMI=body weight/(height)2 in kg/m2. In the World Health Organization guidelines for Asians, individuals with a BMI 23 kg/m2 are classified as overweight and those with a BMI 25 kg/m2 are defined as obese (Fenrick et al., 2000). We recruited case group (n=206) that comprised overweight and obese subjects (BMI≥23 kg/m2) and control subjects (n=152) with normal weights (BMI; 18.0∼22.9 kg/m2) at Kyung Hee University Medical Center and Keimyung University Dongsan Medical Center. All of the subjects were ethnic Koreans. Blood samples were drawn for biochemical measurements, i.e., fasting plasma glucose (FBS), fasted glycated hemoglobin (HbA1c), triglyceride (TG), total cholesterol (TC), high-density lipoprotein cholesterol (HDL-C), and low-density lipoprotein cholesterol (LDL-C). The clinical characteristics of the subjects were summarized (Table 1). DNA was isolated from peripheral blood using the G-DEXTM IIb Genomic DNA Extraction Kit (iNtRON Biotechnology, Seongnam, Korea). All of the subjects gave written informed consent before entering the study. This study was approved by the Institutional Review Board
of Kyung Hee University Medical Center.
2. SNP selection and genotyping
Initially, we selected 55 tagging SNPs within the ETV6 gene in the intron and exon, using the following criteria: (1) consisting only of SNPs in introns (approximately 300 bp from the exon to consider alternative splicing); (2) tagging SNPs selected using the tagging option of the program Tagger (http://www.broad.mit.edu/
mpg/tagger/); (3) known heterozygosity and minor allele frequency (MAF)>0.05; and (4) reported validation (http://www.
hapmap.org). The genotyping was performed using the Affy- metrix Targeted Genotyping Chip array (Affymetrix, CA, USA), according to the manufacturer protocol. This chip uses molecular inversion probe technology with gene chip universal microarrays to provide a method that is capable of analyzing thousands of variants in a single reaction. In brief, DNA was digested with Xba I (New England Biolabs, Beverly, MA), and was subjected to PCR using primers specific to the adaptor sequence (P/N 900409, Affymetrix). PCR products were purified and the fragmented DNA was then end-labeled with biotin using terminal deoxynucleotidyl transferase. Labeled DNA was then hybridized onto the Mapping Array. The hybridized array was washed, stained, and scanned according to the manufacturer's instructions.
The image was analyzed using GCOS software (Affymetrix).
Table 2. Logistic analysis of overweight/obese with ETV6 gene polymorphism in Korean population.
Loci Genotype Overweight/obese (freq.) Control (freq.) Model OR (95% CI) p
rs2723825 C/C 125 (0.59) 115 (0.72) Codominant 0.56 (0.37∼0.84) 0.0048
C/T 82 (0.38) 42 (0.26) Dominant 0.53 (0.34∼0.83) 0.0054
T/T 6 (0.03) 2 (0.01) Recessive 0.44 (0.09∼2.25) 0.3000
rs1781804 C/C 162 (0.78) 101 (0.65) Codominant 1.72 (1.15∼2.58) 0.0081
C/T 40 (0.19) 48 (0.31) Dominant 1.93 (1.20∼3.10) 0.0063
T/T 5 (0.02) 7 (0.04) Recessive 1.80 (0.55∼5.91) 0.3300
rs2710304 C/C 78 (0.36) 68 (0.42) Codominant 0.73 (0.53∼1.00) 0.0500
C/T 101 (0.47) 78 (0.49) Dominant 0.77 (0.50∼1.18) 0.2300
T/T 35 (0.16) 14 (0.09) Recessive 0.48 (0.25∼0.94) 0.0270
rs7296981 C/C 129 (0.60) 74 (0.47) Codominant 1.48 (1.05∼2.07) 0.0230
C/T 73 (0.34) 69 (0.44) Dominant 1.64 (1.08∼2.51) 0.0210
T/T 12 (0.06) 15 (0.09) Recessive 1.55 (0.69∼3.46) 0.2800
Logistic regression models were used for calculating odds ratios (OR) [95% confidence interval (CI)] and corresponding p-values for each SNP site using SNPstats. p-values of codominant, dominant, and recessive models are also given. Age and sex were adjusted by inclusion in logistic analysis as covariates, p-value was significant after correction for Bonferroni (significant: p<0.009).
3. Statistical analysis
For the case-control association study, Hardy-Weinberg equilibrium (HWE) for all SNPs was assessed using SNPstats (Rambaldi et al., 2007). A linkage disequilibrium (LD) block of polymorphisms was tested using Haploview (version 4.12) with Gabriel method (Gabriel et al., 2002; Barrett et al., 2005).
Multiple logistic regression models were calculated for the odds ratio (OR), 95% confidence interval (CI) and corresponding p- values, controlling for age and sex as covariables. We calculated sample powers for the SNPs to confirm the effects. The power of the sample size was calculated using a genetic power calculator (http://pngu.mgh.harvard.edu/~purcell/gpc) (Purcell et al., 2003).
To reduce experimental error, the effective sample size was adjusted (calculated sample size 100/95).
Clinical characteristics were compared between control and overweight/obese subjects, using Student’s unpaired t-test for continuous variables with normal distribution including systolic blood pressure (SBP), diastolic blood pressure (DBP), FBS, HbA1c, TG, TC, HDL-C, and LDL-C. For all of the statistical tests, the level of significance was set at 0.05. Additional statistic analyses using Bonferroni correction were applied to adjust for multiple comparisons.
RESULTS
Table 1 shows the clinical and metabolic characteristics of the overweight/obese control subjects. BMI values were used to categorize 358 subjects into control (BMI; 18∼22.9, n=152) and overweight/obese (BMI≥23, n=206) subjects. The mean value of BMI in the overweight/obese group was significantly higher than that in the control subjects (p<0.001). The levels of SBP, DBP, FBS, TG, TC and, LDL-C in the overweight/obese were higher compared to those of the control subjects (p<0.005).
Also, the level of HDL-C was relatively lower in the over- weight/obese subjects (p<0.001).
We also analyzed for association between each genotype and susceptibility to overweight/obesity by logistic regression analyses, after adjusting for age and sex. The genotype distributions of these fifty five SNPs in the ETV6 were in HWE (p>0.05). Four individual polymorphisms (rs2723825, rs17818048, rs2710304, and rs7296981) were found to be associated with overweight/
obesity (Table 2). Using the Bonferroni method, statistical signi- ficances of two SNPs (rs2723825 and rs17818048) were preserved in the codominant model and dominant model (codominant:
p=0.0048, dominant: p=0.0054; codominant: p=0.0081, domi- nant: p=0.0063, respectively, Table 1).
Through pair-wise linkage analysis between overweight/obese and control group, we have found that seventeen LD blocks
Fig. 1. (A) Gene map in ETV6. Coding exons are marked by shade block, and 5,3-UTRs indicated by white blocks. (B) Haplotypes of ETV6. Only those with frequencies>0.1 including significant SNPs are presented. (C) Linkage disequilibrium coefficients (│D'│and r2) among the ETV6 polymorphism. r2=1 means absolute LDs among polymorphisms. Asterisks (*) indicate significant SNPs in case and control comparison.
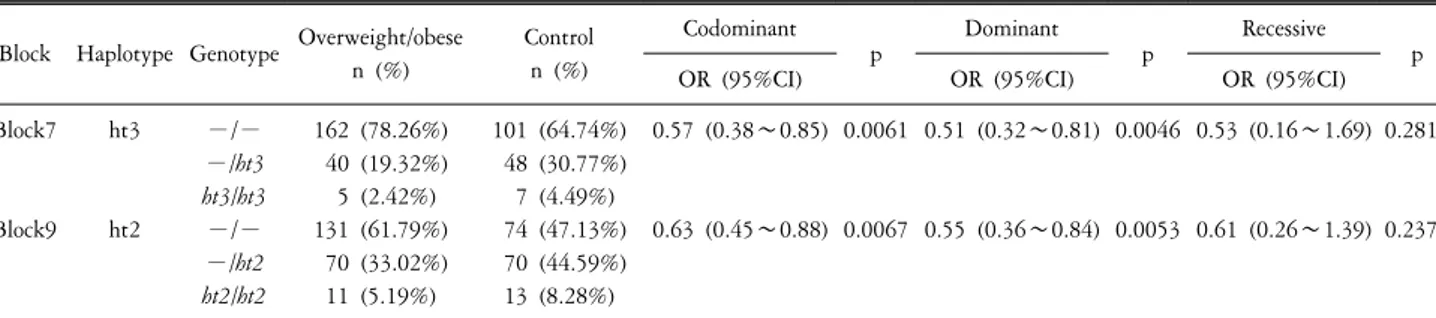
Table 3. Logistic analysis of overweight/obese with haplotype of ETV6 in Korean population.
Block Haplotype Genotype Overweight/obese n (%)
Control n (%)
Codominant
p Dominant
p Recessive
OR (95%CI) OR (95%CI) OR (95%CI) p
Block7
Block9 ht3
ht2
−/−
−/ht3 ht3/ht3
−/−
−/ht2 ht2/ht2
162 (78.26%) 40 (19.32%) 5 (2.42%) 131 (61.79%) 70 (33.02%) 11 (5.19%)
101 (64.74%) 48 (30.77%) 7 (4.49%) 74 (47.13%) 70 (44.59%) 13 (8.28%)
0.57 (0.38∼0.85)
0.63 (0.45∼0.88) 0.0061
0.0067
0.51 (0.32∼0.81)
0.55 (0.36∼0.84) 0.0046
0.0053
0.53 (0.16∼1.69)
0.61 (0.26∼1.39) 0.2818
0.2378
Missing genotype data were omitted for exact haplotype construction. p-values are for regression analyses of three alternative models (codominent, dominant, and recessive models). p-values of haplotype association were calculated using HapAnalyzer.
ETV6 were constructed using Haploview (version 4.12, data not shown) (Fig. 1). Among the haplotypes (ht), ht3 in block7 and ht2 in block9 were used for further analysis because the haplotypes in block7 and block9 were including significant SNPs (rs17818048, rs2710304, and rs7296981, respectively). Haplotypes
with either equivalence with single polymorphism or with frequencies of less than 1% were excluded from further analysis.
In further analysis, we found ht3 (GGG) of block7, consisting rs2724623, rs17818048, and rs2710304, showed susceptibility to the risk of overweight/obesity in the codominant model (p=
0.0061, OR= 0.57, 95% CI=0.38∼0.85) and dominant model (p=0.0046, OR=0.51, 95%CI=0.32∼0.81). Ht2 (CCC) of block9, consisting rs7296981, rs1981440, and rs2710287, were significantly associated with overweight/obese in the codominant model (p=0.0067, OR=0.63, 95%CI=0.45∼0.88) and dominant model (p=0.0053, OR=0.55, 95%CI=0.36∼0.84), respectively.
However, other haplotype in each block showed no significant associations (Table 3).
DISCUSSION
This study shows that the genetic polymorphisms and the haplotype of ETV6 are associated with the overweight/obesity.
Recently, numerous candidate genes were investigated to find out the genetic factors implicated in the pathogenesis of overweight/
obesity (Clément, 2006).
ETV6 is member of the ETS family of transcription factors.
ETS factors bind heterogenous sequences centered around a core GGA sequence and cooperate with other transcription factors to regulate the transcription of a diverse set of genes (Graves et al., 1998). Occurrence of somatic heterozygous mutations of the ETV6 gene in the acute myeloid leukemia (AML) was reported.
Furthermore, defected ETV6 protein expression was appeared in one-third of AML patient (Barjesteh et al., 2005). Overweight/
obesity may be increase the risk of developing leukemia (Larsson et al., 2007). The biologic mechanism underlying the observed association of overweight/obese with increase risk of leukemia is unclear. It could be associated to the decreased immune response and chronic inflammation associated with obesity (Chandra, 1997;
Marti, 2001; Tilg et al., 2006).
In this study, we investigated whether the ETV6 gene polymorphisms are related to overweight/obese subjects by genotyping the fifty five selected SNPs in Korean population.
Our results showed that some SNPs in the ETV6 contribute to overweight/obese in Korean population. In a case-control analysis, of the fifty five SNPs examined in ETV6, four SNPs (rs2723825, rs17818048, rs2710304, and rs7296981) were significantly associated with risk of overweight/obese. Moreover, haplotypes of the ETV6 ht3 in block7 and ht2 in block9 had a significant association with the risk of overweight/obesity. The minor allele of rs2723825 and rs2710304 had susceptibility to the risk of
overweight/obese in the codominant model, dominant model, and recessive model, respectively. Conversely, the minor allele of 1781804 and rs7296981 showed protective effect to overweight/obese in the codominant model, dominant model, respectively. In addition, ht3 of block7 and ht2 of block9 in ETV6 were associated with risk of overweight/obese. There is no report on the association of ETV6 with overweight/obesity. This result is first to show significant association between SNPs in ETV6 and overweight/obese.
In conclusion, this result revealed that SNPs and haplotypes in ETV6 may be contributing to the association overweight/obesity in Korean population, possibly with other genetic and environmental factors. Further work from different populations with larger sample size and corresponding functional evidence, is still needed to confirm the suggestive associations of ETV6 SNPs with overweight/obesity.
ACKNOWLEDGEMENTS
This work was supported by Kyung Hee University Research Year Program (2006).
REFERENCES
Barjesteh van Waalwijk van Doorn-Khosrovani S, Spensberger D, de Knegt Y et al. (2005) Somatic heterozygous mutations in ETV6 (TEL) and frequent absence of ETV6 protein in acute myeloid leukemia. Oncogene. 24:4129-4137.
Barrett JC, Fry B, Maller J et al. (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21:263- 265.
Chandra RK (1997) Nutrition and the immune system: an introduction. Am. J. Clin. Nutr. 66:460S-463S.
Clément K (2006) Genetics of human obesity. C. R. Biol. 329:608- 622.
Fenrick R, Wang L, Nip J et al. (2000) TEL, a putative tumor suppressor, modulates cell growth and cell morphology of ras-transformed cells while repressing the transcription of stro- melysin-1. Mol. Cell. Biol. 20:5828-5839.
Gabriel SB, Schaffner SF, Nguyen H et al. (2002) The structure of haplotype blocks in the human genome. Science 296:2225-2229.
Golub TR, Barker GF, Lovett M et al. (1994) Fusion of PDGF receptor beta to a novel ets-like gene, tel, in chronic myelo- monocytic leukemia with t(5;12) chromosomal translocation. Cell.
= 국문초록 =
Ets variant 6 (ETV6) 유전자는 염색체의 재배열과 관련이 있는 백혈병에 관련이 되었다고 알려져 있다. 이번 연구에 서, 과체중이 백혈병과 과체중/비만에 위험 요소로 작용을 조사하였다. 55개의 single nucleotide polymorphisms (SNPs)을 Affymetrix Genotyping chip array 방식을 이용 분석하였는데, 분석에는 206명의 과체중/비만과 152명의 한국 정상인이 참여되었다. 그리고 통계분석으로는 logistic and multiple regression 방법이 사용되었다. 55개의 polymorphism 중, 4개 (rs2723825, rs17818048, rs2710304 그리고 rs7296981)에서 유의성이 관찰되었다. 그리고 2개의 하플로 타입 중에 블록 7(GGG)과 9(CCC)에서 유의성이 있는 확인할 수 있었다. 유의성을 보였던 블록에는 유의성이 있던 polymorphism이 포함되어 있었다. 우리의 실험 결과에서, ETV6는 한국인의 과체중/비만에 관계가 있다는 것을 밝혀졌다.
중심단어: ETV6, Haplotype, 비만, 단일염기다형성 77:307-316.
Graves BJ, Petersen JM (1998) Specificity within the ets family of transcription factors. Adv. Cancer. Res. 75:1-55.
Larsson SC, Wolk A (2007a) Body mass index and risk of multiple myeloma: a meta-analysis. Int. J. Cancer. 121:2512-2516.
Larsson SC, Wolk A (2007b) Obesity and risk of non-Hodgkin's lymphoma: a meta-analysis. Int. J. Cancer. 121:1564-1570.
Larsson SC, Wolk A (2007c) Overweight and obesity and incidence of leukemia: A meta-analysis of cohort studies. Int. J. Cancer.
15:1418-1421.
Leonhardt M, Hrupka B, Langhans W (1999) New approaches in the pharmacological treatment of obesity. Eur. J. Nutr. 38:1-13.
Marti A, Marcos A, Martinez JA (2001) Obesity and immune function relationships. Obes. Rev. 2:131-140.
Odero MD, Carlson K, Calasanz MJ et al. (2001) Identification of
new translocations involving ETV6 in hematologic malignancies by fluorescence in situ hybridization and spectral karyotyping.
Genes. Chromosomes. Cancer. 31:134-142.
Purcell S, Cherny SS, Sham PC (2003) Genetic Power Calculator:
design of linkage and association genetic mapping studies of complex traits. Bioinformatics 19:149-150.
Rambaldi D, Felice B, Praz V et al. (2007) Splicy: a web-based tool for the prediction of possible alternative splicing events from Affymetrix probeset data. BMC Bioinformatics 8 Suppl 1:S17.
Tilg H, Moschen AR (2006) Adipocytokines: mediators linking adipose tissue, inflammation and immunity. Nature Reviews 6:772-783.
Wlodarska I, Mecucci C, Baens M et al. (1996) ETV6 gene rearrangements in hematopoietic malignant disorders. Leuk.
Lymphoma. 23:287-295.