저작자표시-비영리-변경금지 2.0 대한민국 이용자는 아래의 조건을 따르는 경우에 한하여 자유롭게 l 이 저작물을 복제, 배포, 전송, 전시, 공연 및 방송할 수 있습니다. 다음과 같은 조건을 따라야 합니다: l 귀하는, 이 저작물의 재이용이나 배포의 경우, 이 저작물에 적용된 이용허락조건 을 명확하게 나타내어야 합니다. l 저작권자로부터 별도의 허가를 받으면 이러한 조건들은 적용되지 않습니다. 저작권법에 따른 이용자의 권리는 위의 내용에 의하여 영향을 받지 않습니다. 이것은 이용허락규약(Legal Code)을 이해하기 쉽게 요약한 것입니다. Disclaimer 저작자표시. 귀하는 원저작자를 표시하여야 합니다. 비영리. 귀하는 이 저작물을 영리 목적으로 이용할 수 없습니다. 변경금지. 귀하는 이 저작물을 개작, 변형 또는 가공할 수 없습니다.
Characterization of Genes Specifically
Expressed in Embryonic and Cancer Stem Cells
by
Jeong A Shin
Major in Neuroscience
Department of Biomedical Sciences
The Graduate School, Ajou University
Characterization of Genes Specifically
Expressed in Embryonic and Cancer Stem Cells
by
Jeong A Shin
A Dissertation Submitted to The Graduate School of
Ajou University in Partial Fulfillment of the Requirements
for the Degree of
Master of Neuroscience Sciences
Supervised by
Myung Ae Lee, Ph.D.
Major in Neuroscience
Department of Biomedical Sciences
The Graduate School, Ajou University
This certifies that the dissertation
of Jeong A Shin is approved.
SUPERVISORY COMMITTEE
Myung Ae Lee
Hye Sun Kim
Joon Kyu Lee
The Graduate School, Ajou University
June 22nd, 2012
i
- ABSTRACT -
Characterization of Gene Co-expressed
In Both Embryonic Stem Cells and Cancer Cells
ES cells are derived from the ICM of blastocyst and have ability of self-renewal, unlimited proliferation and pluripotency that differentiate into cells of all three germ layers. The stem cell genes such as Pou5F1 (Oct4, Oct3/4), Sox2, Nanog, LIN28, KLF4, Myc (c-Myc) act alone and interact with each other for mainintaining the stem cell state. These genes have been already studied well. The emergence of new genes and the interaction with new genes can enhance research in the stem cell field. Some studied show that human embryonic stem cell genes are expressed in several cancer cells. Cancer cells or cancer stem cells have the similar characteristics. In order to understand the mechanism of the similar characteristics such as self-renewal, the development potential and the enhancement of proliferative ability, we find OR that is expressed in both stem cells and cancer cells using DDD. We make OR overexpressed in the human neural stem cells with the natural existence of OR expression accessory protein. This OR changed cell cycle indicating that OR acts as a regulatory member of cell cycle control system. This study is the clue to understand the basic mechanism of cell cycle control system of ES cells and cancer cells. Futhermore it can contribute in the field of ES cell and cancer cell research and therapy.
Key word : Embryonic stem cell, cancer stem cell, self-renewal, stemness, cell cycle, olfactry recepter
ii
TABLE OF CONTENTS
ABSTRACT ··· ⅰ TABLE OF CONTENTS ··· ⅱ LIST OF FITURES ··· ⅳ LIST OF TABLES ··· ⅴ ABBREVIATION ··· ⅵ Ⅰ. INTRODUCTION ··· 1Ⅱ. MATERIAL AND METHODS ··· 3
A. Digital differential dispay··· 3
B. Cell culture ··· 3
C. Vector construct for OR overexpression ··· 3
D. Infection ··· 4
E. RNA preparation and Reverse transcription-polymerase chain reaction ··· 4
F. Cell cycle analysis ··· 5
G. Cell proliferation assay··· 6
H. Microarray··· 6
I. Statical Analysis ··· 6
Ⅲ. RESULTS ··· 7
A. Selection of highly expression genes in human embryonic stem cells and cancer cells··· 7
iii
C. Effect of OR on cell cycle progression ··· 11
D. Existence of OR expression accessory gene ··· 12
Ⅳ. DISCUSSION ··· 21
Ⅴ. CONCLUSION ··· 25
REFERENCES ··· 26
iv
LIST OF FIGURES
Fig.1. Searching data for digital differential display ··· 8
Fig.2. Overexpression of OR in F3 cells, HEK293 cells and NIH3T3 ··· 10
Fig.3. OR-infected F3 cells and HEK293 cells increase S phase ··· 12
Fig.4. Increased proliferation of F3 cells and HEK293 cells overexovessing OR ··· 13
Fig.5 Human neural stem cells include other non-OR GPCR ··· 17
Fig.6 Expression of subtype M3R ··· 18
Fig.7 Cell proliferation change by muscarinic receptor agonist and antagonist ··· 19
v
LIST OF TABLE
vi
ABBREVIATION
DDD : digital differential display hNSC : human neural stem cell hESC ; human embryonic stem cell OR : olfactory receptor
PI : propidium iodide
1
Ⅰ
. INTRODUCTION
Embryonic stem (ES) cells, derived from the inner cell mass (ICM) of blastocysts, possess three characteristic features: First, the ability to renew themselves, second, unlimited proliferation, and third, pluripotency to differentiate into a variety of tissues (Thomson et al, 1998; Jiang et al, 2007). These unique characteristics of ES cells have led to expectations that human ES cells might be useful to understand disease mechanism and injuries such as spinal cord injury, to develope for drug and transplantation medicine, and to adjust cell replacement therapies (Takahashi et al, 2007; Ittai et al 2008).
Cancer cell is also an important research object because it cannot be conquered yet since we have been studying it for long time. Cancer cells grow out of control, develop abnormal size and shapes, ignore their typical boundaries inside the body, and can ultimately spread or metastasize to other organs and tissues. Cancers are indeed extremely diverse and heterogeneous (Miller, 1981). The specific stem properties are relavant to some forms of human cancer. Tumor-initiating cells have been identified in cancers of the hematopoietic system, brain, and breast. These cells have the capacity for self-renewal, the potential to develop into any cell in the tumor population, and the proliferative ability to expand continuously the population of malignant cells. Accordingly, the properties of tumor-initiating cells are almost similar to the three features that define normal stem cells. Malignant cells with these functional properties are called “cancer stem cells” (Jordan et al, 2006).
2
Consistent with this notion, many studies have revealed that stem cells including embryonic stem cells have very similar characterisitics to cancer cells. Specifically, Oct4 and Nanog genes, which are important for human ESC stemness, are expressed in several cancer cells (Dick, 2008; Ezeh et al, 2005), and Myc, another ES-specific gene, is known to act as oncogene in human lung cancer cell line (Little, 1983). Even though stem cell and cancer cell share similar properities, the different properties exist. ES cell is affected by extrinsic cue (e.g., growth factors, ligands secreted by or attached to other cells, physical stresses, and other external influences) in deterministic manner and intrinsic cell mechanisms (e.g., gene expression) in a stochastic manner. These factors determine stem cell fate transition. However, cancer cell have intrinsic resistance (e.g., chemotherapeutic agents) (Hiddessen, 2005; Takashita, 2000).
"Stemness" is core stem cell properity that underline self-renewal in both stem cells and cancer cells (Cai et al, 2004; Ramalho-Santos et al, 2002). To understand molecular background of stemness, we performed Digital differential display (DDD) to find out the genes which are expressed not in differentiated tissues, but in both ES and cancer cells.
3
Ⅱ. MATERIALS AND METHODS
A. Digital differential display
In a typical digital differential display experiment, the user must select specific tissue libraries to be assigned to each pool. These pools were then compared. DDD compares the Expressed Sequence Tags (ESTs) constituents of various tissue types, depending on selected libraries thereby determining the relative representation of each sequence in the libraries being compared. The DDD output was in the form of a web file that had links to Unigene clusters that correspond to the EST's which were differentially expressed between the two tissues.
B. Cell culture
F3 (immotalized human neural stem cell, NCSs), HEK293 (human embryonic kidney cell) and NIH3T3 (mouse fibroblasts cells) were maintained and passaged on uncoated culture dishes in Dulbecco's modified medium (DMEM, Sigma aldrich) with 10% Fetal bovine serum (FBS, Hyclone), 10μg/ml penicillin-streptomycin (Gibco). 293GPG cells were maintained and passaged on uncoated culture dishes in Dulbecco's modified medium (DMEM, Sigma aldrich) with 10% Fetal bovine serum (FBS, Hyclone), 10μg/ml penicillin-streptomycin (Gibco), 1μg/ml tetracycline, 2μg/ml puromycin and 300μg/ml G418.
C. Vector construct for OR overexpression
pCXbsr vector and pCL Ampho vector (helper construct for retrovirus) were kindly provided by Dr Inoue in Shiga university of medical science, of Japan. Both pCXbsr and
4
pCMV6-XL5-OR56B1 (ORIGENE) were digested by EcoRⅠ and did ligation at 4℃. The constructs were confirmed by digestion with EcoRⅠ and AccⅠ.
D. Infection
pCXbsr-OR56B1 vector and pCL Ampho vector were transfected to 293GPG cells using the lipofectamine and plus reagent according to the manufacturer's protocol (Invitrogen). 2μg pCXbsr vector or 2μg pCXbsr-OR56B1 vector and 2μg pCL Ampho vector were used for transfection when the cells were 80~90% confluent. 293GPG cells incubated
at 37℃ in a CO2 incubator for 2 days. The cells were collected on 100mm dish and 293GPG
cells were incubated at 37℃ in a CO2 incubator for 2 days. F3 cells, HEK293 cells and
NIIH3T3 cells treated with DEAE-DEXTRAN (Sigma aldrich) in DMEM were infected by
filtered 293GPG media soup and incubated in the CO2 incubator with 37℃ for 2 days. Then
media was exchanged with media containing blasticidin S (Invitrogen) and selected for 1-2weeks.
E. RNA preparation and reverse transcription-polymerase chain reaction
Total RNA was isolated using TRIzol (Invitrogen) according to the manufacturer's protocol. cDNA was synthesized using 2μg total RNA in the presence of SuperscriptⅡ (Invitrogen) and oligo(dT) (Invitrogen). The reaction mixture was incubated at 42℃ for 50mins followed by incubation at 72℃ for 15mins. 150ng of cDNA was then used as
template in 50μl PCR product containing 10x PCR buffer, 1.5mM MgCl2, 0.2mM
5
(Invitrogen). The PCR condition of OR56B1 was as follow :30 cycles as 94℃ for 45 seconds, 56.7℃ for 45 seconds, 72℃ for 45 seconds and a final extension for 10 minutes at 72℃. The primer sequences were as follow : ATG AAT CAT ATG TCT GCA TCT CTC-3' and 5'-TAG ATC ACT CCC CAT TCC AAG C-3'. The PCR condition of CHRM3 was as follow : 30 cycles as 94℃ for 45 seconds, 62.6℃ for 45 seconds, 72℃ for 45 seconds and a final extension for 10 minutes at 72℃. The primer sequences were as follow 5'-CCA AAG AGC TTG CTG GCC TG-3' and 5'-CCA CGC TCT TCT GGG CCT G-3'. PCR products were separated using 1.2% agarose gel with TAE buffer.
F. Cell cycle analysis
NIH3T3 cells transfected with none, pCX vector and pCX-OR56B1 vector respectively were
seeded at a concentration of 3X105 cells in 3ml DMEM with 10% FBS and 1%
penicillin/streptomycin in 60mm dishes. F3 cells and HEK293 cells were transfected with none, pCX vector and pCX-OR56B1 vector and were seeded at a concentration of 8X105 cells in 8ml DMEM with 10% FBS and 10μg/ml penicillin-streptomycin in 100mm dishes. Cells were washed with PBS and then fixed with ice-cold 70% ethanol for at least 30 mins at 4℃. Fixed cells were washed with PBS and incubated in PBS containing 0.2mg/ml RNase A (Sigma-Aldrich) for 15 min at 37°C. 40μg/ml propidium iodide (PI; Sigma-Aldrich) was added to the samples and incubated for 30 min at 37°C. Samples were subjected to FACS Aria Ⅲ (BD Biosciences, Mountain View, CA) analysis using FACs Diva II software.
6 G. Cell proliferation assay
NIH3T3 cells infected with none, pCX vector and pCX-OR vector were seeded in triplicate at a density of 10000cells in 35mm dishes in 4ml media volume respectively. F3 cells and HEK293 cells infected with none, pCX vector and pCX-OR vector were seeded in triplicate at a density of 40000cells on 35mm dishes in 4ml media volume respectively. Cells were detached by trypsinization and dyed by 0.4% tryphan blue solution, and Cell number was measured on a CountessTM Cell Counting slide (Invoitrogen) by CountessTM automated cell
counter (Invoitrogen) for 5 days.
H. Microarray Analysis
Total RNA was isolated from samples of human neural stem cell line, A4 and F3. After cDNA synthesis and label for biotin by T7 Megascript kit (Ambion, Austin, TX), hybridization was predormed using GeneChip Fluidixs station protocol EukGE_WS1 (Affymetrix; Santa Clara, DA) and GeneChip3.0 software (Affymetrix) was used for scan and analysis.
I. Statistical Analysis
Statistics were applied with one- way ANOVA (SPSS 16, IBM, Somers, NY) along with a post hoc Fisher’s significant difference test (SPSS 16). ). α ,p < 0.05 ; *, p < 0.005 ; **, p < 0.0005 ; ***, p < 0.000005.
7
Ⅲ
. RESULTS
A. Selection of highly expressied genes in human embryonic stem cells and cancer cells
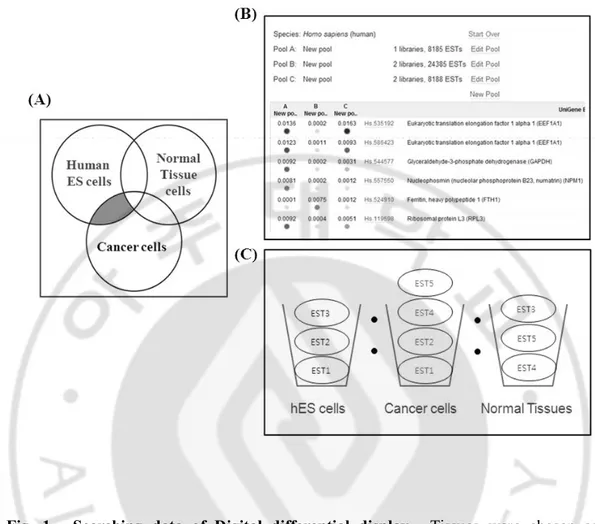
As mentioned above, human embryonic stem cells maintain their specific characterization by several genes. Genes such as Oct4, Nanog, Sox2 and Klf4 are already known well. Beside these genes, we searched for new gene as a regulator of the cell cycle control system of stem cells and cancer stem cells using digital differential display. We selected specific tissue libraries such as hES cell, cancer cell and normal tissue to be assigned to each pool. These pools were then compared. DDD compares the Expressed Sequence Tags (ESTs) constituents of various tissue types, depending on selected libraries thereby determining the relative representation of each sequence in the libraries being compared. The DDD output was in the form of a web file that had links to Unigene clusters that correspond to the EST's which were differentially expressed between the two tissues (Fig.1). Selected genes were not only expressed in human embryonic stem cells but also expressed in cancer cells other than normal cells (Fig.1A). Finally we chose one gene that is olfactory receptor because OR strongly expressed in hES cells (no data).
8
Fig. 1. Searching data of Digital differential display. Tissues were chosen as an interesting target. The system returns the number of UniGenes of different specificities in each tissue. After clicking a number on the last page, the system returns target tissues UniGenes matching the chosen criteria. After page users can set parameters for comparisons with secondary tissues
9 B. Overexpression of Olfactory receptor
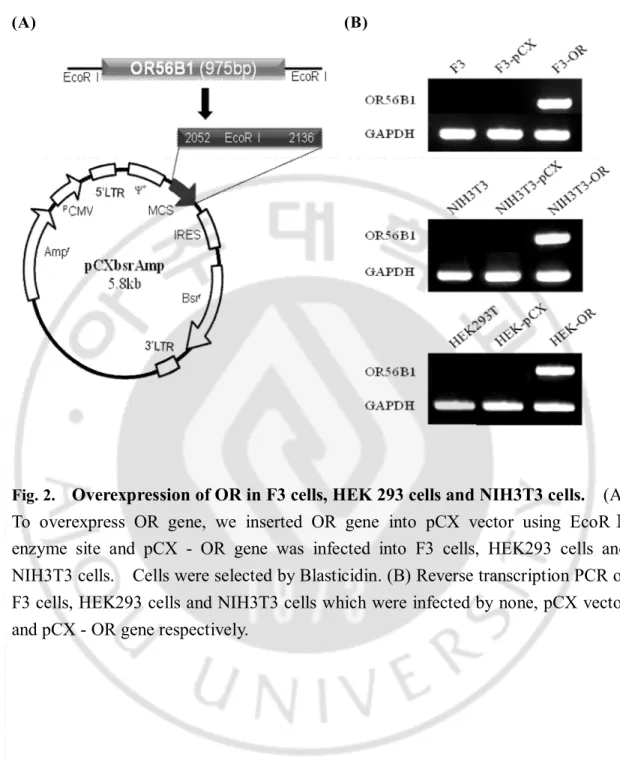
To overexpress OR gene, we inserted OR gene into pCX vector using EcoRⅠenzyme site and this construct was confirmed by digestion reaction (Fig.2A). pCX retrovirus vector carrying OR gene (pCX-OR) and pCLAmpo vector (helper vector for retrovirus) were transfected into 293GPG cells. 293GPG cells made the intact viruses for infection. Finally F3 cells (Human neuronal stem cell line), HEK293 cells (human embryonic kidney cell line) and NIH3T3 cells (Mouse embryonic fibroblast cell line) were infected with pCX-OR and followed by selection with Blasticidin. pCX vector was used as control.
Reverse transcription PCR of F3 cells, HEK293 cells and NIH3T3 cells infected by none, pCX vector and pCX-OR gene was performed respectively. None and pCX vector were control. OR gene expression was extremly high in OR infected cells, but F3 cells, HEK293 cells and NIH3T3 did not express OR gene (Fig.2B).
10 (A) (B)
Fig. 2.
Overexpression of OR in F3 cells, HEK 293 cells and NIH3T3 cells. (A)
To overexpress OR gene, we inserted OR gene into pCX vector using EcoRⅠ
enzyme site and pCX - OR gene was infected into F3 cells, HEK293 cells and
NIH3T3 cells. Cells were selected by Blasticidin. (B) Reverse transcription PCR of
F3 cells, HEK293 cells and NIH3T3 cells which were infected by none, pCX vector
and pCX - OR gene respectively.
11 C. Effect of OR on cell cycle progression
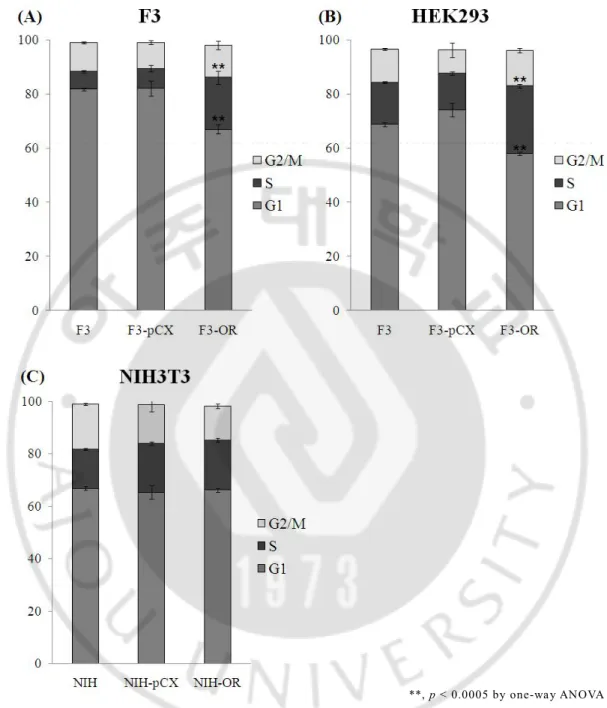
In order to know the effector of OR on cell cycle progression, we carried flow cytometry analysis on PI-labeled F3 cells infected with none, pCX vector and pCX-OR vector respectively. F3 cells and HEK293 cells which were infected by pCX-OR vector were dynamically changed showing decrease G1 phase and increase S phase (Fig.3A and B).
In the same way, we performed flow cytometry analysis of NIH3T3 cells infected with none, pCX vector and pCX-OR vector respectively. Cells were dyed by PI. No valuable change was observed (Fig.3C).
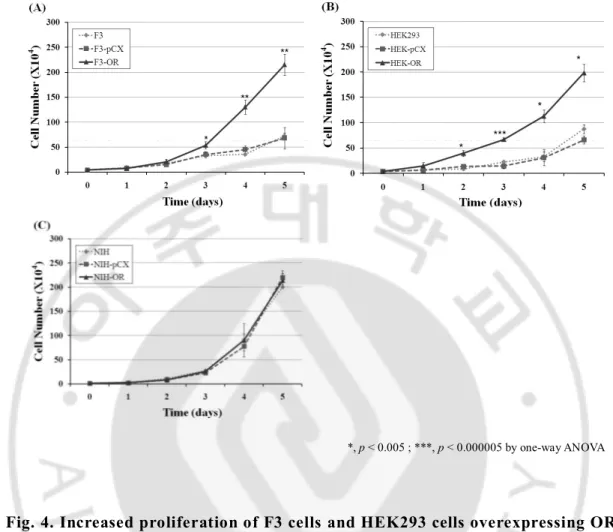
To clarify that change of cell cycle by OR actually effects the cell proliferation, we counted cell number for 5 days. NIH3T3 cells infected with none, pCX vector and pCX-OR vector, and F3 cells and HEK293 cells infected with none, pCX vector and pCX-OR vector were seeded at a density of 10000cells and 40000cells respectively. Proliferation curve showed that proliferation curve of F3 cells and HEK293 cells infected with pCX-OR was extremely changed comparing other cells infected with none, and pCX vector (Fig.4A and B). But the data is shown no change in NIH3T3 cells with none, pCX vector and pCX-OR vector (Fig.4C).
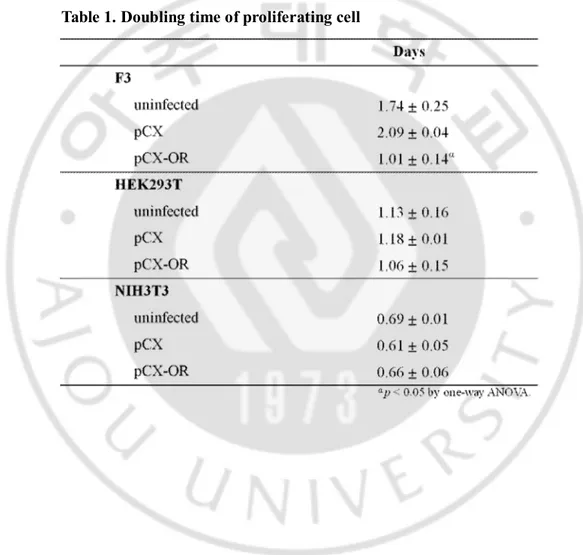
The doubling time of these cells (Table1) was calculated for 3 to 5 days. HEK293 cells was calculated for 1 to 5 days. The data is shown in Fig. 3. Doubling time of F3 cells and HEK293 cells infected with OR were decreased as compared to F3 cells infected with none, pCX vector. NIH3T3 cells infected with none, pCX vector and OR were similar time. Doubling time of F3 cell and infected with OR was ~50% shorter than F3 cells infected with non and pCX vector. HEK293 cells show the same result.
12
**, p < 0.0005 by one-way ANOVA
Fig. 3. OR-infected F3 cells and HEK293 cells increase S phase. (A), (B) and (C) Cell cycle analysis of F3 cells, HEK293T cells and NIH3T3 cells with none, pCX vector and pCX-OR vector respectively. Experiment was performed by flow cytometry on PI-labeled cells (n=3). **, p < 0.0005 by one-way ANOVA with a post hoc Fisher’s least significant difference test. Error bars represent the standard deviation
13
*, p < 0.005 ; ***, p < 0.000005 by one-way ANOVA
Fig. 4. Increased proliferation of F3 cells and HEK293 cells overexpressing OR.. (A), (B) and (C) F3 cells, HEK293T cells and NIH3T3 cells infected with none,
pCX vector and pCX-OR vector are measured by CountessTM automated cell
counter respectively (n=3). *, p < 0.005 ; ***, p < 0.000005 by one-way ANOVA with a post hoc Fisher’s least significant difference test. Error bars represent the standard deviation.
14
Table 1. Doubling time of proliferating cell. The doubling time of F3 cells and HEK293T cells with OR was decreased compared with F3 cells infected with none, pCX vector. NIH3T3 cells infected with none, pCX vector and OR have the same doubling time. Results of F3 cells, HEK293 cells and NIH3T3 cells were obtained from days 1 to 5 (for Fig. 4 data, n = 3). Significance was determined by one-way ANOVA with a post hoc Fisher’s least significant difference test.
15 D. Existence of OR expression accessory gene
According to the hypothesis of M3 muscarinic acetylcholine receptor (M3R) modulates Odorant receptor signaling, we presume human neuronal stem cells (F3, A4 and F5 cells) were naturally expressed functional factor which modulate OR signaling such as M3R.
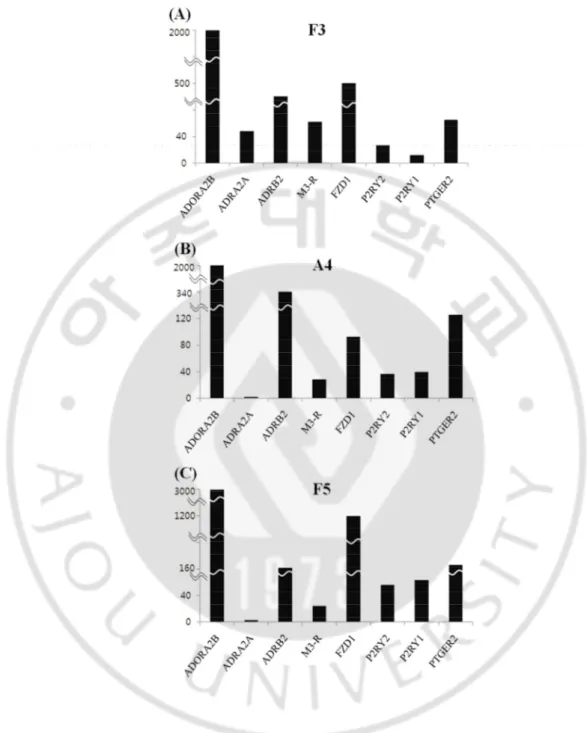
There is possibility that other non-OR GPCR also may modulate OR signaling. Microarray data shows non-OR GPCRs are existed in human neural stem cells; Adenisine A2B receptor, Alpha-2A adrenergic receptor, Beta-2 adrenergic receptor,
Muscarinic Acetylcholine receptor3, Frizzled receptor 1, Purinergic P2Y receptor, Purinergic P1Y receptor and Prostaglandin E2 receptor (Fig. 5).
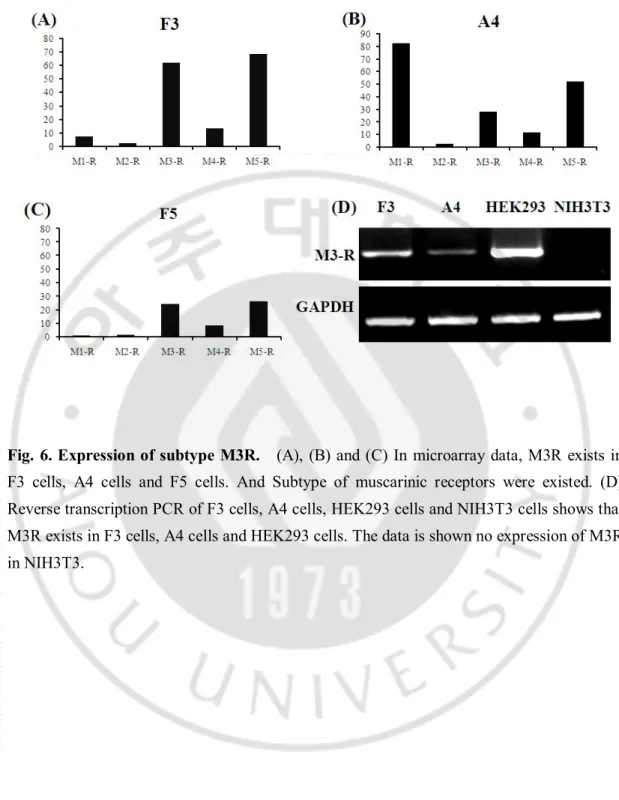
In the microarray, other muscarinic receptor subtype including subtype 3 are naturally expressed in human neural stem cell (Fig.6). Reverse transcription PCR result shows actual existence of M3R in human neuronal stem cell and human embryonic kidney cell. The data is shown no expression in differentiated cell (Fig.6D).
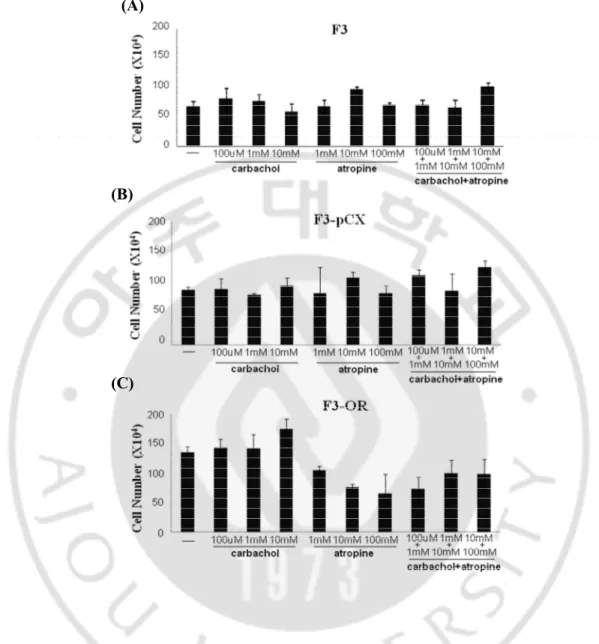
To convince muscarinic receptor help cell progression with OR, we analysis cell proliferation change with muscarinic receptor agonist and antagonist. We treat carbachol and atropine as agonist and antagonist respectively in dose dependent manner (Fig.8). The data of F3-OR cells shows that cell proliferation increase by carbachol in dose dependent manner and decrease by atropine in dose dependent manner. This is a consistent pattern comparing other cells.
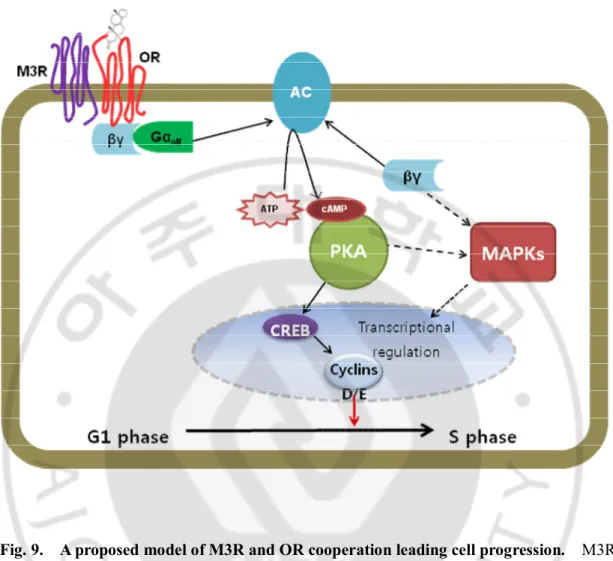
Model of of M3 muscarinic acetylcholine receptor (M3R) and OR interaction shows how the M3-R affect OR function and what might be the molecular
16
mechanism of this interaction. M3R modulates mammalian odorant receptor signaling through the interaction between M3R and olfactory receptor. Binding
odorant and OR activates an odorant-specific G-protein (Golf) that, in turn, activates
an adenylate cyclase, resulting in the generation of cyclic AMP (cAMP). Accumulation of cAMP leads to the activation of the transcription factor. The resulting effect on cell cycle progression is dependent on a number of factors, including the concentration of cAMP generated. PKA can also regulate, positively or negatively, other mitogenic pathways, particularly those leading to the activation of MAPKs.
17
Fig. 5. Human neural stem cells include other non-OR GPCR. In microarray data, Neural stem cell line, (A) F3 cells, (B) A4 cells and (C) F5 cells indicate the expression of non-olfactory receptor GPCR.
18
Fig. 6. Expression of subtype M3R. (A), (B) and (C) In microarray data, M3R exists in F3 cells, A4 cells and F5 cells. And Subtype of muscarinic receptors were existed. (D) Reverse transcription PCR of F3 cells, A4 cells, HEK293 cells and NIH3T3 cells shows that M3R exists in F3 cells, A4 cells and HEK293 cells. The data is shown no expression of M3R in NIH3T3.
19 (A)
(B)
(C)
Fig. 7. Cell proliferation change by muscarinic receptor agonist and antagonist. (A) F3 cells , (B) F3-PCX cells and (C) F3-OR cells are incubated for 5 days in the presence of the muscarinic agonist carbachol (100µM, 1mM, 10mM), the muscarinic antagonist atropine (1mM, 10mM, 100mM), and cabachol and atropine. Cells are measured by CountessTM
20
Fig. 9. A proposed model of M3R and OR cooperation leading cell progression. M3R and OR heteromers may couple with G proteins more efficiently than do monomeric receptors. And G-protein of OR and M3R effect cell proliferation. Futhermore this cell proliferation ability is included in ES cell properities.
21
Ⅳ
. DISCUSSION
Stem cells have three distinctive properties. These are self renewal, the capability to develop into multiple lineages and the potential to extensively proliferate. Stem cell is unique because of these three properties (Jordan et al, 2006).
Several genes that modulate stem cell-like state have been studied. These stem cell maker genes such as Nanog, Oct4, Sox2, cMyc, Lin28 and KLF4 are highly expressed in human embryonic stem cells and make somatic cell induce stem cell. Futhermore Oct4 and nanog are expressed in cancer cell lines (Thomson et al, 1998; Yu et al, 2007; Dick, 2008; Ezeh et al, 2005).
Accordingly the gene research related stemness is an important field to understand the mechanism of induced stem cell, cancer cell and cancer stem cell.
First of all, In order to find the genes which are expressed in both stem cell and cancer cell except normal tissue cell, we used Digital Differential Display (DDD). Digital Differential Display (DDD) is a powerful web-based bio-informatic tool to identify differential gene expression. DDD uses the EST profiles of cDNA libraries which are represented in the NCBI Uni-Gene database (Murray et al, 2007).
We selected tissue libraries which are to be assinged to each pool. DDD compared assignments of ESTs from different libraries, or pools of libraries to a specific UniGene cluster. The DDD output linked to Unigene clusters that correspond to the ESTs that were differentially expressed between the two tissue.
Among 11 genes, we chose one, an olfactory receptor gene, which confirmed gene expression in human ES cell compared with other tissue and neural stem cell.
22
Olfactory receptor (OR) belongs to the G-protein-coupled receptor (GPCR) superfamily and are seven transmembrane domain (7TM) proteins (Bakalyar and Reed, 1990; Buck and Axel, 1991). Approximately 950 ORs are distributed in ~100 locations throughout the genome. About 72% are pseudogenes and ~350 intact and potentially functional genes (Rouquier et al, 1998; Gaillard et al, 2004).
OR can be divided into two classes depending on ligand. One is classⅠ(fish-liked receptor) that is ~10% of that of mammalian species, another is classⅡ (tetrapod specific receptor) is the majority of mammalian OR genes. The current hypothesis is that ligand of class I ORs could be hydrophobic volatile odorants, whereas ligand of class II ORs would be hydrophobic compounds (Glusman et al, 2001; Gaillard et al, 2004).
The structure of OR is GPCRs which feature seven hydophobic stretches, which including the transmembrane domains, a potential disulfide bond between the highly conserved cysteines in extracellular loops 1 and 2, a conserved NXS/T consensus for glycosylation in the N-terminal region, several potential phosphorylation sites in intracellular regions and numerous conserved short sequences. (Strader et al, 1994; Gat et al, 1994)
The specific characteristics to ORs is an unusually long second extracellular loop, two conserved cysteines in this loop and conserved amino acid motifs which indicate a vertebrate 7TM. The transmembrane domains 3, 4 and 5 are hypervariable. They make the odorant binding pocket. (Gaillard et al, 2004).
OR expresses in olfactory sensory neurons (OSNs) of the olfactory epithelium located in the posterior upper part of the nasal cavity. When odorants activate OR, it transduces intracellular signals by coupling to GTP - binding protein.
23
OR signal transduction from chemical information to electrical impulses arise via a G-protein-coupled activation of an adenylyl cyclase. It leads to increase cyclic AMP (cAMP) and to open consequently the cyclic nucleotide-activated, nonselective cation channels. The influx of cations makes the olfactory neuron depolarize and olfactory neuron result in action potentials. OR signal is integrated at the cortex level to result in the sensation of smell. This is original pathway by olfactory receptor (Bakalyar and Reed, 1990; Gaillard et al, 2004).
These specialized isoforms of Gαolf, adenylyl cyclase type III and the cyclic nucleotide-activated channel in the olfactory cilia suggest the importance of this pathway. Moreover, Brunet et al suggested that the cAMP cascade is dominant when odorant signals are transmitted in the olfactory neurons in gene knockout studies (Brunet et al, 1996), but the role of an inositol-1,4,5-triphosphate (IP3)-mediated pathway is not clearly revealed (Kaur et al, 2001).
In order to have overexpression, we performed digestion of olfactory receptor gene and pCXbsrAmpo retrovirus vector by EcoRⅠ and then ligation. It is confirmed by restriction enzyme reaction.
pCXbsrAmp was backbone of an olfactory gene and pCL Ampho was a helper vector for retrovirus. Therefore, Both of them were co-transfected to 293GPG cell that allowed for the production of high titer amphotropic retrovirus.
After incubation, we collected soup and NIH3T3 cells (mouse embryonic fibroblast cells) were infected. But there was no change in the flow cytometry result. In the same way, F3 cells (human neural stem cells) and HEK293 cells (human embryonic kidney cells) were infected. Flow cytometry result showed cell cycle phases were extremly changed. G1
24
phase was decrease but S phase was increase. This olfactory receptor gene effect as cell cycle regulator in case of F3 cells and HEK293 cells.
OR is difficult to express in heterologous cells. When OR express in common cell culture systems, the OR proteins are detected intracellularly. It means OR protein has very little localization to the plasma membrane (McClintock, 2003).
In order to enchance transporting and expression of OR, OR need accessary proteins such as GPCR, transporting and expression enchacing protein, and Heat shock protein. The non-OR GPCR usually make heterodimer with OR to localize properly at the plasma membrane (Prinster et al, 2005). For instance the β2-adrenergic receptor, the P2Y1, P2T2
and adenosine A2a receptors increase amount of the OR-M71 (Bush and Hall, 2008). M3R, one of the non-OR GPCR, has different function that might lead to change in OR conformation or ligand-induced changes in OR conformation that improve ligand binding or that M3-R and OR heteromers may couple with G proteins more efficiently than do monomeric receptors. (Li and Matsunami, 2011). But this hypothesis is not clear. The microarray and RT-PCR results showed that F3 cells and HEK293 cells have M3R, P2RY1 and P2RY2 exist in quantity in microarray results. (Von et al, 2006) We also confirm the agonist and antagonist of M3R influence cell proliferation.
Through this fact, we expect when OR overexpress in the F3 cells and HEK293 cells, cell proliferation is extremely increased indicating accessory proteins existence in these cells to help in OR expression and enhance OR signaling.
But we do not yet know the molecular mechanism that OR effect cell cycle change. And we need to clarify the roles and interaction of OR and M3R. Therefore we focus to reveal questions in the futher study.
25
Ⅴ
. CONCLUSION
In this study, we found new gene expressed in both stem cell and cancer cell using DDD. This gene make human neural stem cell cycle change with accessory proteins which are naturally expressed in human neural stem cell and related cell proliferation. As a result, The cells overexpressing OR decreased G1 phase and increase S phase. Human embryonic kidney cells also show the same change. But the change is not shown in mouse embryonic fibroblast cells.
The results of cell cycle change indicate a possibility that OR can participate in cell cycle control system as a regulator in stem cell helping OR accessory protein.
These results provide a new idea for the regulation mechanism of embryonic stem cell, cancer cell and cancer stem cell.
26
REFERENCES
1. Bakalyar HA, Reed RR. Identification of a specialized adenylyl cyclase that may mediate odorant detection. Science 7;250(4986):1403-6, 1990
2. Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet 40(5): 499–507, 2008
3. Brunet LJ, Gold GH, Ngai J. General anosmia caused by a targeted disruption of the mouse olfactory cyclic nucleotide-gated cation channel. Neuron 17(4):681-93, 1996
4. Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell 5;65(1):175-87, 1991
5. Bush CF, Hall RA. Olfactory receptor trafficking to the plasma membrane. Cell Mol Life Sci 65(15):2289-95, 2008
6. Cai J, Weiss ML, Rao MS. In search of “"stemness”". Exp Hematol 32(7):585-98, 2004
7. Dick JE. Stem cell concepts renew cancer research. Blood 15;112(13):4793-807, 2008
8. Ezeh UI, Turek PJ, Reijo RA, Clark AT. Human embryonic stem cell genes OCT4, NANOG, STELLAR, and GDF3 are expressed in both seminoma and breast carcinoma. Cancer 15;104(10):2255-65, 2005
27
9. Fuchs T, Glusman G, Horn-Saban S, Lancet D, Pilpel Y. The human olfactory subgenome: from sequence to structure and evolution. Hum Genet 108(1):1-13, 2001
10. Gaillard I, Rouquier S, Giorgi D. Olfactory receptors. Cell Mol Life Sci 1(4):456-69, 2004
11. Gat U, Nekrasova E, Lancet D, Natochin M. Olfactory receptor proteins. Expression, characterization and partial purification. Eur J Biochem 1;225(3):1157-68, 1994
12. Glusman G, Yanai I, Rubin I, Lancet D. The complete human olfactory subgenome. Genome Res 11(5):685-702, 2001
13. Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced Pluripotent Stem Cell Lines Derived from Human Somatic. CellsScience 21;318(5858):1917-20, 2007
14. Jiang Y, Jahagirdar BN, Reinhardt RL, Schwartz RE, Keene CD, Ortiz-Gonzalez XR, Reyes M, Lenvik T, Lund T, Blackstad M, Du J, Aldrich S, Lisberg A, Low WC, Largaespada DA, Verfaillie CM. Pluripotency of mesenchymal stem cells derived from adult marrow. Nature 4;418(6893):41-9. Epub 2002 Jun 20, 2002
15. Jordan CT, Guzman ML, Noble M. Cancer Stem Cells. N Engl J Med 21;355(12):1253-61, 2006
28
16. Kaur R, Zhu XO, Moorhouse AJ, Barry PH. IP3-Gated Channels and their Occurrence Relative to CNG Channels in the Soma and Dendritic Knob of Rat Olfactory Receptor Neurons. J Membr Biol 15;181(2):91-105, 2001
17. Li YR, Matsunami H. Activation state of the M3 muscarinic acetylcholine receptor modulates mammalian odorant receptor signaling. Sci Signal 11;4(155):ra1, 2011
18. McClintock TS, Sammeta N. Trafficking prerogatives of olfactory receptors. Neuroreport 26;14(12):1547-52, 2003
19. Murray D, Doran P, MacMathuna P, Moss AC. In silico gene expression analysis –an overview. Mol Cancer 7;6:50, 2007
20. Okita K, Ichisaka T, Yamanaka S. Generation of germline - competent induced pluripotent stem cells. Nature 19;448(7151):313-7, 2007
21. Prinster SC, Hague C, Hall RA. Heterodimerization of g protein-coupled receptors: specificity and functional significance. Pharmacol Rev 57(3):289-98, 2005
22. Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA. “Stemness”: Transcriptional Profiling of Embryonic and Adult Stem Cells. Science 18;298(5593):597-600, 2002
23. Rouquier S, Taviaux S, Trask BJ, Brand-Arpon V, van den Engh G, Demaille J, Giorgi D. Distribution of olfactory receptor genes in the human genome. Nat Genet 18(3):243-50, 1998
29
24. Strader CD, Fong TM, Tota MR, Underwood D, Dixon RA. Structure and function of G protein-coupled receptors. Annu Rev Biochem 63:101-32, 1994
25. Synnergren J, Giesler TL, Adak S, Tandon R, Noaksson K, Lindahl A, Nilsson P, Nelson D, Olsson B, Englund MC, Abbot S, Sartipy P. Differentiating human embryonic stem cells express a unique housekeeping gene signature. Stem Cells 25(2):473-80, 2007
26. Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 30;131(5):861-72, 2007
27. Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science 6;282(5391):1145-7, 1998
28. Von Dannecker LE, Mercadante AF, Malnic B. Ric-8B promotes functional expression of odorant receptors. Proc Natl Acad Sci U S A 13;103(24):9310-4, 2006
30 -국문요약-
배아줄기세포와
암세포에서
공통적으로
발현되는 유전자의 특성 분석
아주대학교 대학원 의생명과학과 신 정 아 (지도교수: 이 명 애) 포유류의 배반포에서 유래한 배아줄기세포의 세가지 큰 특징은 자가증식 할 수 있고, 체세포나 생식세포 등 모든 형태로 분화가 가능하다는 점과 끊임없이 분열 할 수 있는 증식능력이다. 이러한 배아줄기세포의 능력은 Pou5F1 (Oct4, Oct3/4), Sox2, Nanog, LIN28, KLF4, Myc (c-Myc)과 같은 유전자들이 스스로 혹은 상호 작용을 통해 유지된다. 이러한 주요 유전자는 이미 많은 연구가 이루어 지고 있는 바이며, 이 연구분야를 확장시키기 위한 새로운 유전자의 등장과 다른 유전자와의 상호작용에 대한 연구가 기대 되어지는 바이다. 최근엔 이 유전자들 중 Oct4, Nanog 가 암세포에서도 발견 되어진다는 연구가 있다. 암세포에서 활발히 연구 되어지고 있는 대상인 암줄기세포는 줄기세포와 같은 특징을 공유한다. 줄기세포와 암줄기세포의 중복되는 유전자를 연구함으로써, 배아줄기세포뿐 아니라 암세포에 대한 공통된 특징이나 각각의 이해를 돕는 새로운 메커니즘을 제시한다.31
우리는 Digital differential display (DDD)를 통해서 인간배아 줄기세포와 여러 가지 다양한 암세포들에서 공통적으로 발현하는 유전자 중 가능성이 높은 후각수용기의 한 유전자를 선택하였다. 이 유전자를 과발현시키기 위해 바이러스 벡터에 주입하였으며, 바이러스를 이용한 형질전환방법을 이용하였다. 이를 Mouse embryonic fibroblast cell, Human neural stem cell 과 Human embryonic kidney cell 에 주입하여, 세포주기를 측정하였다. 오직 Human neural stem cell 과 Human embryonic kidney 에서만 세포주기가 변화되는데, 이는 후각수용기 유전자의 작용을 증폭시키는 보조 단백질인 Muscarinic receptor3 가 세포 자체에서 발현되어지고 있기 때문에, 후각 수용기 유전자가 오직 Human neural stem cell 과 Human embryonic kidney 에서만 세포주기에 영향을 미치고 있음을 알 수 있다.
인간배아줄기세포와 암세포에 공통적으로 발현되면서, 세포주기조절능력에 영향을 미치는 새로운 유전자에 대한 연구는 인간배아줄기와 암세포를 동시에 연구할 수 있는 단서를 제공하며, 이 세포들에 관한 연구와 치료에 있어 큰 공헌을 할 것이라 기대된다. 핵심어 : 배아줄기세포, 암줄기세포, 다능성, 자가증식, 줄기성, 세포주기, 후각수용기