Functional characterization of gibberellin signaling-related genes in <i>Panax ginseng</i>
전체 글
수치
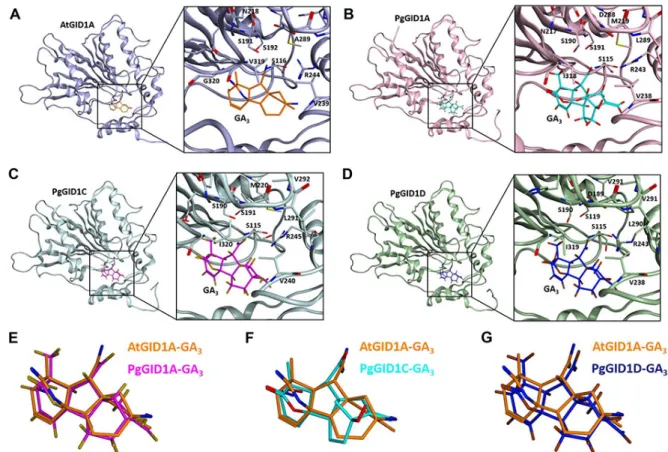
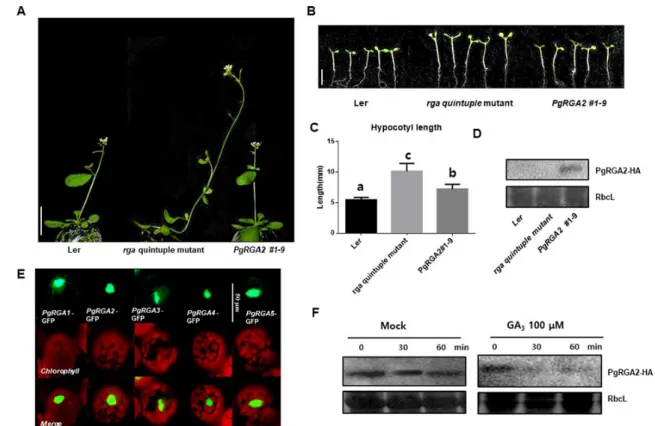
관련 문서
Basic aspects of AUTOSAR architecture and methodology Safety mechanisms supported by AUTOSAR.. Technical safety concepts supported by AUTOSAR Relationship to ISO
1 John Owen, Justification by Faith Alone, in The Works of John Owen, ed. John Bolt, trans. Scott Clark, "Do This and Live: Christ's Active Obedience as the
As a result of the simple analysis, we found that variables related to drinking problems of college students were religion, family, residence, parents'
As a result of the study, it was confirmed that only human resource competency had a positive(+) effect on the intercompany cooperation method of IT SMEs,
Skin transcriptomes analysis from atopic dermatitis group showed 14 keratinocyte differentiation related genes, 10 negative regulation of peptidase related
Phylogenetic analysis of Orientia tsutsugamushi strains based on the sequence homologies of 56-kDa type-specific antigen genes. FEMS
The analysis was done on the school section that needs to be improved first and is the most basic out of the different issued related to the high school equalization such
As a result of this study, it was confirmed that the possibility of valuable resources recovery was positive when a combined process of froth