Korean Journal of Microbiology (2019) Vol. 55, No. 4, pp. 445-447 pISSN 0440-2413
DOI https://doi.org/10.7845/kjm.2019.9108 eISSN 2383-9902
Copyright ⓒ 2019, The Microbiological Society of Korea
Complete genome sequence of clinical strain, Streptococcus pneumoniae 521
Hayoung Lee
1,2and Seung Il Kim
1,2*
1
Research center for Bioconvergence Analysis, Korea Basic Science Institute (KBSI), Cheongju 28119, Republic of Korea
2
Department of Bio-Analytical Science, University of Science and Technology, Daejeon 34113, Republic of Korea
임상 균주 Streptococcus pneumoniae 521의 유전체 염기서열 해독
이하영
1,2・ 김승일
1,2*
1
한국기초과학지원연구원 바이오융합연구부,
2과학기술연합대학원대학교 생물분석과학
(Received September 18, 2019; Revised December 3, 2019; Accepted December 3, 2019)
*For correspondence. E-mail: ksi@kbsi.re.kr;
Tel.: +82-43-240-5422; Fax: +82-43-240-5416
Streptococcus pneumoniae is the main cause of community- acquired pneumonia (CAP). Here we present the complete annotated genome sequence and antibiotic-resistance genes of S. pneumoniae 521, which was isolated from a patient. The genome comprises a single circular 2,041,494 bp chromosome with a G + C content of 39.7%. The genome contains 144 antibiotic-resistance genes. This genomic data will facilitate a better understanding of the transfer mechanisms of antibiotic resistance and the development of novel drugs.
Keywords: Streptococcus pneumoniae, clinical strain, complete genome sequence, pacbio, pneumonia
Streptococcus pneumoniae (S. pneumoniae) is the major causative bacterium of community-acquired pneumonia (CAP) (Mandell et al., 2007). Despite the fact that many clinical cases of S. pneumoniae still remained susceptible to clinically used antibiotics, many multidrug resistant strains have frequently emerged recently (Cho et al., 2014). To prevent the transmission of multidrug resistance, the selection of an appropriate antibiotic treatment is important. In this regard, the genome information of clinical strains isolated from domestic patients is very useful
because genomic variation can reflect differential antibiotic resistance characteristics. In this report, we present the complete genome sequence of the clinical strain; S. pneumoniae 521 which was isolated from the respiratory track of a patient in Seoul, Korea.
S. pneumoniae 521 was cultivated anaerobically in Tryptic Soy Broth at 37.5°C in 5% CO
2for 7~8 h. The bacterial genomic DNA was prepared as previously described (Ro et al., 2018). Genome sequencing of S. pneumoniae 521 was performed at Macrogen, Inc. using the PacBio single molecule real-time (SMRT) platforms. In total, 159,522 reads were obtained and de novo assembled using Unicycler v0.4.8-beta (Wick et al., 2017), resulting in a single circular chromosome (2,041,494 bp) with no plasmids.
Average Nucleotide and Amino Acid Identity (ANI/AAI) distances were calculated according to the MiGA workflow (Rodriguez et al., 2018). Gene prediction was carried out using the NCBI Prokaryotic Genome Annotation Pipeline 4.2 (Tatusova et al., 2016). Virulent and antibiotic-resistance genes were searched against a comprehensive antibiotic resistance database (CARD) (Jia et al., 2017). Resistance islands were predicted using IslandViewer 4 (Bertelli et al., 2017; Abraham et al., 2018).
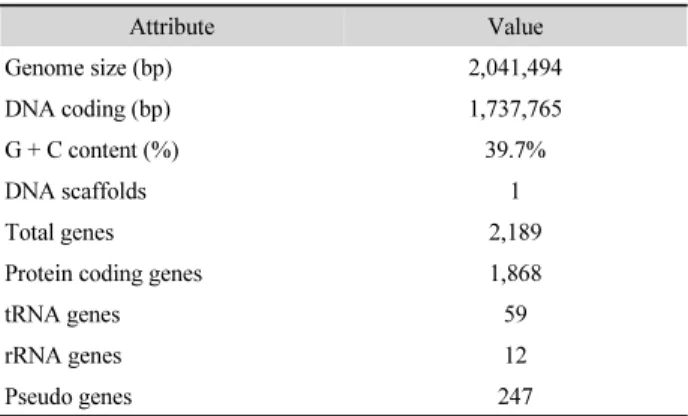
The genome properties of S. pneumoniae 521 are summarized
446
∙ Lee and Kim미생물학회지 제55권 제4호
Table 1. Genome statistics of S. pneumoniae 521
Attribute Value
Genome size (bp) 2,041,494
DNA coding (bp) 1,737,765
G + C content (%) 39.7%
DNA scaffolds 1
Total genes 2,189
Protein coding genes 1,868
tRNA genes 59
rRNA genes 12
Pseudo genes 247
in Table 1. The complete genome size is 2,041,494 bp with a G + C content of 39.7%. The closest relative strain was S.
pneumoniae R6 (NC_003098, 99.98% ANI, 99.89% AAI). As a result of the gene prediction process, this genome was found to contains 2,189 CDSs, 59 tRNA, and 12 rRNA genes. We found 144 antibiotic-resistance related genes using CARD.
However, only five genes [mefE, pmrA, RlmA(II), patA, and patB] were valid using the strict cutoff criterion. These genes are categorized according to their function, such as an efflux pump conferring antibiotic resistance (mefE, pmrA, patA, and patB), and antibiotic target alteration [RlmA(II)], suggesting the potent mechanism of antibiotic resistance that bacteria have. In addition, 36 antibiotic-resistant or virulence-related genes, including mefE, were located in the genomic island (57,752~94,436 bp) which were identified using IslandViewer 4. Interestingly, these regions show high homology levels in some S. pneumoniae (strain R6, R6CIB17, NCTC13276, NCTC7466, D39, and D39V) and a Streptococcus milleri (strain NCTC10708) suggesting a horizontal transfer from common ancestors (Coffey et al., 1991). Taken together, we expect that the availability of the complete genome sequence for S.
pneumoniae 521 will facilitate further study of pneumococcal diseases and their genomic characterizations.
Nucleotide sequence accession number
The Streptococcus pneumoniae 521 is available at KCTC 43179. This complete genome sequence was deposited at NCBI GenBank under the accession CP036529. The version described in this paper is version CP036529.1
적 요
Streptococcus pneumoniae는 지역획득폐렴의 주요 원인 균 이다. 우리는 환자로부터 분리된 S. pneumoniae 521의 염기서 열을 분석하고 항생제 내성 유전자를 확인하였다. 이 균의 유 전체는 G + C 비율이 39.7%인, 2,041,494 bp 크기의 염기서열 로 이루어져있다. 이 유전체에서 총 144개의 항생제 내성 유전 자를 확인하였다. 이러한 유전체 정보가 항생제 내성 전달 기 전에 대한 이해나 신약 개발에 유용할 것으로 기대된다.
Acknowledgements
This work was supported by UST Young Scientist Research Program through the University of Science and Technology (No. 2019YS05).
References
Abraham S, Kirkwood RN, Laird T, Saputra S, Mitchell T, Singh M, Linn B, Abraham RJ, Pang S, Gordon DM, et al. 2018.
Dissemination and persistence of extended-spectrum cephalosporin- resistance encoding IncI1-blaCTXM-1 plasmid among Escherichia coli in pigs. ISME J. 12, 2352–2362.
Bertelli C, Laird MR, Williams KP, Simon Fraser University Research Computing G, Lau BY, Hoad G, Winsor GL, and Brinkman FSL. 2017. Islandviewer 4: Expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 45, W30– W35.
Cho SY, Baek JY, Kang CI, Kim SH, Ha YE, Chung DR, Lee NY, Peck KR, and Song JH. 2014. Extensively drug-resistant Streptococcus pneumoniae, South Korea, 2011-2012. Emerg. Infect. Dis. 20, 869–871.
Coffey TJ, Dowson CG, Daniels M, Zhou J, Martin C, Spratt BG, and Musser JM. 1991. Horizontal transfer of multiple penicillin‐ binding protein genes, and capsular biosynthetic genes, in natural populations of Streptococcus pneumoniae. Mol. Microbiol. 5, 2255–2260.
Jia B, Raphenya AR, Alcock B, Waglechner N, Guo P, Tsang KK, Lago BA, Dave BM, Pereira S, Sharma AN, et al. 2017. Card 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 45, D566– D573.
Mandell LA, Wunderink RG, Anzueto A, Bartlett JG, Campbell GD, Dean NC, Dowell SF, File TM Jr, Musher DM, Niederman MS, et al. 2007. Infectious diseases society of America/American Thoracic Society consensus guidelines on the management of
Complete genome of S. pneumoniae 521∙
447
Korean Journal of Microbiology, Vol. 55, No. 4
community-acquired pneumonia in adults. Clin. Infect. Dis. 44Suppl 2, S27–72.
Ro HJ, Lee H, Park EC, Lee CS, Kim SI, and Jun S. 2018.
Ultrastructural visualization of Orientia tsutsugamushi in biopsied eschars and monocytes from scrub typhus patients in South Korea. Sci. Rep. 8, 17373.
Rodriguez RL, Gunturu S, Harvey WT, Rossello-Mora R, Tiedje JM, Cole JR, and Konstantinidis KT. 2018. The Microbial Genomes Atlas (MiGA) webserver: taxonomic and gene diversity analysis
of archaea and bacteria at the whole genome level. Nucleic Acids Res. 46, W282–W288.
Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, and Ostell J. 2016. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44, 6614–6624.
Wick RR, Judd LM, Gorrie CL, and Holt KE. 2017. Unicycler:
Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13, e1005595.