Kor. J. Hort. Sci. Technol. 30(5):549-556, 2012 DOI http://dx.doi.org/10.7235/hort.2012.11058
An Efficient Identification of 68 Apple Cultivars Using a Cultivar Identification Diagram (CID) Strategy and RAPD Markers
Wenyan Wang1, Kun Wang2, Fengzhi Liu2, and Jinggui Fang1*
1College of Horticulture, Nanjing Agricultural University, Nanjing 210095, P. R. China
2Research Institute of Pomology, Chinese Academy of Agricultural Sciences, Xingcheng 125100, P. R. China
Abstract. The study aimed to establish an efficient tool for cultivar identification and characterization being the first steps of apple introduction and improvement program. We utilized a method to efficiently record DNA molecular fingerprints of plant individuals genotyped by RAPD, which could be used as efficient reference information for quick plant identification. Ten of sixty 11-mer primers were screened to identify the 68 apple genotypes which could be distinguished by a combination of several primers. All cultivars were easily identified by the corresponding primers marked on the cultivar identification diagram (CID). The results indicated that the CID strategy developed and employed in the apple cultivar identification could be vital in the utilization of DNA marker in other plants as well as the development of the apple industry.
Additional key words: fruit industry, longer primer, molecular marker, polymorphic band
*Corresponding author: fanggg@njau.edu.cn
※ Received 21 June 2011; Revised 3 July 2012; Accepted 12 July 2012. We are thankful for funding support from the NCET Program of China (grant no. NCET-08-0796) and the Fundamental Research Funds for the Central Universities (No. KYJ200909).
Introduction
Apple (Malus domestica) is one of the most economically important woody plants cultivated for its value as a fruit crop as well as for its ornamental beauty (Young et al., 2007).
Most apple cultivars are diploid (2n = 34) while some of the main cultivars are triploid (2n = 3x = 51). China is a major producer and supplier of apples to Asia and most growth in the apple trade in Asia is attributed to imports from China. This represents a very important aspect in the fruit industry, particularly since the sale of fruit trees and planting of orchards represent major investments of time and money. Since cultivar identification and characterization are the first steps of any fruit or other crop introduction and improvement program, rapid identification and characterization of these cultivars would provide valuable information for introduction and genetic improvement of apple cultivars.
With the role of DNA fingerprinting as a powerful tool currently applied widely in forensic science, it can also be equally useful in the genotyping of plants. However, the powerful DNA markers available for plant identification have not made fruit crop variety identification efficient, recordable, and as a easy task as desired in practice, fruits and other crop scientists owning to the potential of this technique in
commercial production are facing an awkward situation. The main reason for this awkward situation is dearth of analysis strategies for DNA fingerprints. The currently popular analysis techniques for DNA banding patterns known as cluster analyses cannot be applied efficiently in cultivar or species separation.
This has made practical application of DNA marker in crop and seed identification unfeasible. Various DNA-based markers have recently been developed and used for studies on genetic diversity, fingerprinting, and cultivar origins (Cheng and Huang, 2009; D’Onofrio et al., 2009; Elidemir and Uzun, 2009;
Melgarejo et al., 2009; Papp et al., 2010). Randomly amplified polymorphic DNA (RAPD) markers used in apple before have proven to be a reliable marker system for genetic fingerprinting and also in determining the genetic relationships among germplasm collections. RAPD markers have the advantage of being simple, able to detect relatively small amounts of genetic variation and do not need prior information on the genome. However, it should be pointed out that RAPDs do not give information about the genome. The technique has already been successfully applied to estimate genetic relationships in apricot (Marinello et al., 2002), mulberry (Vijayan, 2004), grape (Benjak et al., 2005), figs (Sadder and Ateyyeh, 2006), litchi (Ding et al., 2000), cherry (Demirsoy et al., 2008), Indian cashew (Archak et al., 2003), longan (Yonemoto et
Table 1. Name, origin and source of the materials used in the experiment (Li et al., 1980; Xin and Xiang, 1998).
No. Cultivar Origin Source
1 ‘Northern Spy’ America Seedling
2 ‘Mutsu’ Japan ‘Golden Delicious’ × ‘Indo’
3 ‘New Jonagold’ Japan ‘Jonagold’ bud sport
4 ‘Stark Spur Supreme’ ‘Red Red America ‘Red Delicious’ bud sport
5 ‘Alps Otome’ Japan ‘Kitakami’ × (‘Tsugarall’ × ‘Summer Pearmain’)
6 ‘Sheng Fang Fu’ Japan ‘Fuji’ bud sport
7 ‘Ji Guan’ China, Liaoning Seedling
8 ‘Xiu Shui Guo Guang’ China, Shandong ‘Ralls Janet’ Seedling
9 ‘Shinsekai’ Japan ‘Fuji’ × ‘Akagi’
10 ‘Hatsuaki’ Japan ‘Jonathan’ × ‘Golden Delicious’
11 ‘Ralls Janet’ America Seedling
12 ‘Lobo’ Canada ‘McIntosh’ seedling
13 ‘Teng Mu 1’ America *
14 ‘Early McIntosh’ America ‘Yellow Transparent’ × ‘McIntosh’
15 ‘Cui Qiu’ China, Liaoning ‘Golden Delicious’ × ‘Yellow Transparent’
16 ‘Chi Cheng’ China, Gansu Seedling
17 ‘Hong Rou Ping Guo’ China, Xinjiang Seedling
18 ‘Aki 5’ Japan ‘Fuji’bud sport
19 ‘Hong Wang Jiang’ China ‘Fuji’ bud sport
20 ‘Miyasaki’ Japan ‘Fuji’ bud sport
21 ‘Summer Pearmain’ America Seedling
22 ‘Senshu’ Japan ‘Toko’ × ‘Fuji’
23 ‘Vista Bella’ America Hybridization
24 ‘Mac Spur’ England ‘McIntosh’ bud sport
25 ‘Orin’ Japan ‘Golden Delicious’ × ‘Indo’
26 ‘Jin Hong’ China, Jilin ‘Golden Delicious’ × ‘Hong tai yang’
27 ‘Ruby Red Spur’ America ‘Starkrimson’ bud sport
28 ‘Well Spur Delicious’ America ‘Fuji’ bud sport
29 ‘Melrose’ America ‘Delicious’ × ‘Jonathan’
30 ‘Starking Red’ America ‘Delicious’ bud sport
31 ‘Aki 2’ Japan ‘Fuji’bud sport
32 ‘Golden Grimer’ America Seedling
33 ‘Qunfu 1’ Japan ‘Fuji’bud sport
34 ‘Nagafu 7’ Japan ‘Fuji’ bud sport
35 ‘Golden Delicious’ America ‘Golden Grimer’ × ‘Golden Reinette’
36 ‘Qiu Fu 1 Hao’ China, Liaoning *
37 ‘Qiufu 1’ Japan ‘Fuji’ bud sport
38 ‘Ying Qiu’ China, Liaoning ‘Jonathan’ × ‘Summer Pearmain’
39 ‘Black Ber Davis’ America ‘Ber Davis’ bud sport
40 ‘Starkrimson’ America ‘Starking Red’ bud sport
41 ‘Giant Jenifon’ China ‘Ralls Janet’ bud sport
42 ‘Close’ America *
43 ‘Cox’s Orange Pippin’ England Hybridization
al., 2006), olive (Belaj et al., 2003) and pear (Stark-Urnau, 2002b; Lee et al., 2004). Among many markers available, RAPD markers are useful for cultivar analysis with important advantages like simplicity, efficiency, relative ease of execution and non-requirement of any previous sequence information (Williams et al., 1990). Optimization of the RAPD technique by choosing 11 mer primers and strict screening PCR annealing temperature before it is employed in fingerprinting plants, can make RAPD a preferable technique for use in plant cultivar identification.
In this study, we use a Cultivar Identification Diagram (CID) which could be utilized as a source of reference
information for 68 apple cultivars identification. The CID shows the separation of 68 selected apple cultivars, based on RAPD banding patterns and can definitely be of great service to the apple industry worldwide.
Materials and Methods
Plant Materials
Young leaves of 68 selected apple cultivars were collected from the Research Institute of Pomology of Chinese Academy of Agricultural Sciences at Xingcheng. The additional information of these cultivars is shown in Table 1.
Table 2. Ten primers were used for the separation of the 68 apple genotypes.
Primer Nucleotide sequence (5’–3’)
Annealing temperature(°C)
Y-3 GTTTCGCTCCA 44.7
Y-5 GTTTCGCTCCG 43.7
Y-6 GTTTCGCTCCC 41.7
Y-9 CTGCTGGGACA 41.7
Y-10 CTGCTGGGACT 42.8
Y-21 GGACCCAACCA 43.7
Y-23 GGACCCAACCG 42.8
Y-47 ACGACCGACAG 44.4
Y-51 TGGTGGCGTTA 43.7
Y-57 ACCCCCGACTA 44.8
Table 1. Countinued.
No. Cultivar Origin Source
44 ‘Han Fu’ China, Shenyang ‘Tohoku’ × ‘Fuji’
45 ‘Fu Shuai’ China, Zhengzhou ‘Early McIntosh’ בGolden delicious’
46 ‘Smith Cider’ America Seedling
47 ‘Jonagold’ America ‘Golden Delicious’ × ‘Jonathan’
48 ‘Discovery’ England Seedling
49 ‘Yellow Transparent’ Russia *
50 ‘Jerseymac’ America ‘NJ24’ × ‘Julyred’
51 ‘Megum’ Japan ‘Ralls Janet’ × ‘Jonathan’
52 ‘Qin Guan’ China, Shanxi ‘Golden Delicious’ × ‘Ji Guan’
53 ‘Sweet Mcintosh’ America ‘Mcintosh’ × ‘Lawver’
54 ‘Silver Spur Red’ America ‘Hi Early’ bud sport
55 ‘Jonathan’ America Seedling
56 ‘Ruby Spur’ America ‘Starkrimson’ bud sport
57 ‘Stark Jumbo’ America ‘Spokane Beauty’ bud sport
58 ‘Bi Hong’ China *
59 ‘Hardispur Delicious’ America *
60 ‘Red Spur’ America ‘Starking Red’ bud sport
61 ‘Tian Huang Kui’ China, Liaoning ‘Summer Pearmain’ × ‘Yellow Transparent’
62 ‘Zao Hong Xing’ China, Shandong ‘Red delicious’ bud sport
63 ‘Liao Fu’ China, Liaoning ‘Lodi’ × ‘Summer Pearmain’
64 ‘Tsugaru’ Japan ‘Golden Delicious’ hybridization
65 ‘Fu Jin’ China, Liaoning ‘Delicious’ × ‘Yellow Transparent’
66 ‘McIntosh’ Canada Seedling
67 ‘Gala’ New Zealand ‘Kidd’s orange Red’ × ‘Golden Delicious’
68 ‘Golden Spur’ America ‘Golden Delicious’ bud sport
*Indicates unknown origin and source of the cultivar.
Genomic DNA Extraction
Total genomic DNA of each genotype was extracted from young leaves using the modified cetyl trimethyl ammonium bromide method (Bousquet et al., 1990; Murray and Thompson, 1980). The extracted DNA was diluted to a final concentration of 30 ng・μL-1 with1 × TE buffer and stored at -20°C before used.
Primers Screening and RAPD Analysis
For RAPD reactions, 60 primers (Yu et al., 2009) were initially tested with a few genotypes and only those primers resulting in clear unambiguous banding patterns with all genotypes tested were selected for use with the full set of genotypes. Consequently, 10 primers (Table 2) that showed well-resolved and reproducible bands were selected to assay all genotypes, while the others were discarded. Reaction solutions consisted of 2.0 μL 10 × buffer, 1.2 μL MgCl2
(25 mM), 1.6 μL dNTP (2.5 mM), 1.6 μL primer (1.0 μM), 0.1 μL rTaq Polymerase Dynazyme (5U/ μL) and 1 μL of genomic DNA, making a total volume of 20 μL. Amplification reactions were performed based on the standard protocol of Williams et al. (1990) with minor modifications. The PCR was carried out in a Autorisierter Thermocycler (Eppendorf, Hamburg, Germany), programmed as follows: an initial pre-denaturation step for 5 min at 94°C; then 42 cycles each
consisting of a denaturation step for 30 s, an annealing step for 1 min at annealing temperature (Table 2) and an extension step for 2 min at 72°C. Amplification was terminated by a final extension of in 72°C for 10 min. After amplification, the amplified DNA fragments were separated by gel electrophoresis in 1.3% agarose (w/v) (Fig. 1) in 1 × TAE (0.04M Tris-acetate, 0.001M EDTA pH 8.0) buffer at 100 V. The gels were stained with 0.5 μg・mL-1 of ethidium bromide and visualized under ultraviolet light for polymorphic bands among the cultivars.
In order to have reproducible, accurate and clear banding
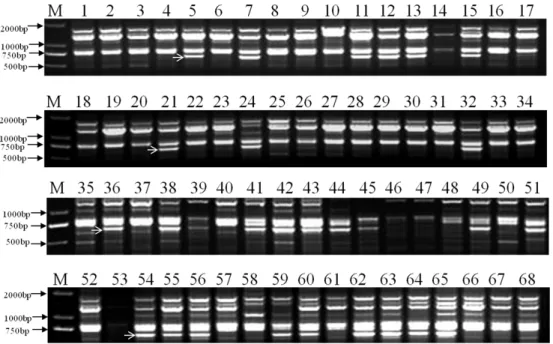
Fig. 1. DNA banding patterns of 68 apple cultivars amplified by primer Y23: M: DL2000 plus DNA ladders; 1-68: Accession numbers of apple cultivars listed in Table 1 are same as those in the Figures. The approximate location of a band for separating is indicated by an arrow.
Fig. 2. The DNA banding patterns of 17 apple cultivars in a group separated using primer Y6: M: DL2000 plus DNA ladders;
the bands pointed with larger arrows were employed in separating the cultivars. The approximate location of a band for separating is indicated by an arrow.
patterns, all amplifications were repeated separately at least thrice.
Data Analysis
Only clear unambiguous bands in the photographic prints of gels were chosen and scored for cultivar identification.
Where some cultivars had a specific band in the fingerprint generated from one primer, they could be separated singly, and those cultivars sharing the same banding pattern were separated and clustered into the same sub-group. Based on this strategy, all the apple cultivars were gradually and completely separated from one another as more primers were employed.
Test of Utilization and Workability of CID in Cultivar Identification
‘Northern Spy’, ‘Mutsu’, ‘New Jonagold’, and ‘Stark Spur Supreme Red Delicious’; ‘Xiu Shui Guo Guang’, and ‘Jin Hong’; ‘Shinsekai’ and ‘Golden Spur’ representing three groups of apple cultivars, randomly chosen from the inter- and intra-groups, were used to verify the utilization and
workability of the diagram showing the separation of the 68 cultivars. The three groups of cultivars were marked “A”,
“B”, and “C” (Fig. 4), and the corresponding primers for use in separation of cultivars in each group were picked out from the CID.
Results
Cultivar Identification
To establish a stable and optimistic RAPD system with high reproducibility, longer primers (11 mer) were employed and the annealing temperatures for each primer were screened based on the quality and reproducibility of banding patterns.
The primers were randomly screened from a stock of 60 11-mer primers, and once an optimistic primer that could produce reproducible polymorphic bands was screened, it was further utilized in identification of the apple cultivars.
By the time the 11th primer was screened and utilized (Table 2), all the 68 apple cultivars could be successfully identified.
An example of the RAPD patterns using primer Y23 in the study was represented in Fig. 1 Electrophoresis results show
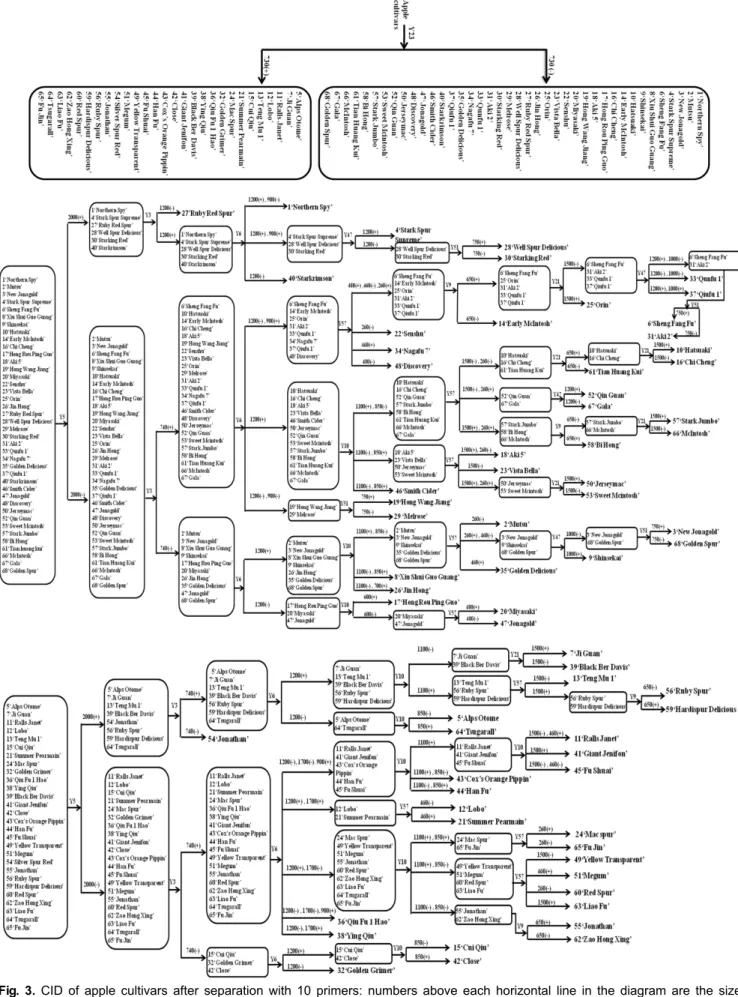
Fig. 3. CID of apple cultivars after separation with 10 primers: numbers above each horizontal line in the diagram are the size of the polymorphic bands in bp used to separate the cultivars following the line; (+) or (-) means presence or absence of the polymorphic band respectively; the cultivar names in bold fonts are those that were separated.
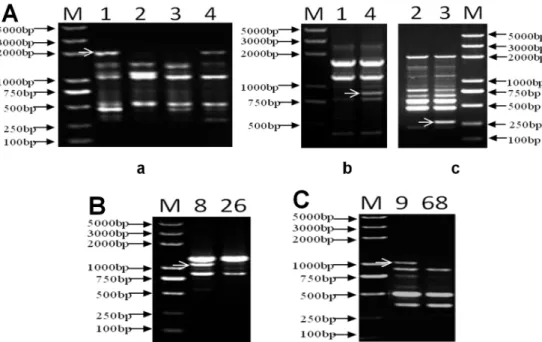
Fig. 4. Result on verification of two cultivars randomly selected by the corresponding primers: M: DL2000 plus marker; “A” is fingerprint obtained with three primers used to separate the first group of selected cultivars denoted “a”, “b”, and “c” and obtained using primers Y5, Y6, and Y57, respectively; “B” is the fingerprint obtained with primer Y10 used to separate the ‘Xiu Shui Guo Guang’and ‘Jin Hong’; “C” is the DNA fingerprint obtained with primer Y47 used to separate the group ‘Shinsekai’ and
‘Golden Spur’. The approximate location of a band for separating is indicated by an arrow.
that primer Y23 generated uniform, clear, and reproducible band patterns in 28 apple cultivars assigned with lane number codes of 5, 7, 11, 12, 13, 15, 21, 24, 32, 36, 38, 39, 41, 42, 43, 44, 45, 49, 51, 54, 55, 56, 59, 60, 62, 63, 64, and 65 (The lane numbers correspond to the cultivar names as shown in Table 1). This group of 28 apple cultivars was easily differentiated from the other 40 cultivars by the presence or absence of a distinct 730 bp band, thus causing the target 68 cultivars to be separated into two groups. The other 9 primers (Table 2) were, step by step, screened and chosen to differentiate the apple cultivars till full separation was achieved as shown in Fig. 3. The second primer (Y5) used could further separate the two groups of cultivars earlier identified by primer Y23 into several smaller groups. After doing these, other primers were chosen to differentiate the several subgroups of apple cultivars respectively. For example, the group comprising of 17 cultivars namely ‘Ralls Janet’,
‘Lobo’, ‘Summer Pearmain’, ‘Mac Spur’ ,‘Qiu Fu 1 Hao’,
‘Ying Qiu’, ‘Giant Jenifon’, ‘Cox’s Orange Pippin’, ‘Han Fu’, ‘Fu Shuai’, ‘Yellow Transparent’, ‘Megumi’, ‘Jonathan’,
‘Red Spur’, ‘Zao Hong Xing’, ‘Liao Fu’, and ‘Fu Jin’, could be further separated into smaller groups or single cultivars e.g ‘Qiu Fu 1 hao’ and ‘Ying Qiu’ with the band sizes of about 1700 bp, 1200 bp, and 900 bp by primer Y6 (Fig. 2).
The apple cultivars could still be separated into many more individual cultivars or groups and systematically following this method and utilizing 10 primers, all the 68 apple cultivars were eventually differentiated from each other (Fig. 3).
Test of the Utilization and Workability of the Diagram in Cultivar Identification
An important aim of this study was to find a strategy of using RAPD markers to distinguish 68 apple cultivars as reported in most of earlier studies focusing on the utilization of DNA marker in identification of plant cultivars. However, there was also a greater need to generate a referable apple CID that could be used in practical identification of some of these cultivars in the future for the nursery industry as well as in cultivar-right-protection. With these two aims in mind, it was critical to verify the utilization, workability, and efficiency of the diagram in cultivar identification. To undertake this, three groups of cultivars comprising of ‘Northern Spy’, ‘Mutsu’, ‘New Jonagold’, and ‘Stark Spur Supreme Red Delicious’; ‘Xiu Shui Guo Guang’, and ‘Jin Hong’;
‘Shinsekai’ and ‘Golden Spur’, which were randomly selected from the inter-and intra-groups in the CID, were used for the verification exercise. Based on the location of these cultivars in the CID, it was easy to find the primer to use in separating them, whereby primers Y5, Y6, Y57, Y10, and Y47 were as projected in the CID used to effectively separate the three groups of cultivars. It was thus clear that PCR verification results agree with those anticipated in the CID with all randomly selected cultivars being separated accordingly. The first group according to Fig. 4A in this study could be validated with three primers namely Y5, Y6, and Y57. The PCR results showed that four apple cultivars
could first be separated into two groups by primer Y5 with a band of about 2000 bp. One group made of ‘Northern Spy’ and ‘Stark Spur Supreme Red’, can be further disjoined using primer Y6 with a 900 bp band. The other group of
‘‘Mutsu’ and ‘New Jonagold’ could then be divided by use of primer Y57 with a band of 260 bp. ‘Xiu Shui Guo Guang’
and ‘Jin Hong’ could be separated with a specific band of about 1100 bp of primer Y10 as in the CID. Another group made up of ‘Shinsekai’ and ‘Golden Spur’ was divided by primer Y47 with the band of about 1000 bp. Following this procedure, all the three groups were successfully identified with suitable primer combinations. This test therefore proves the availability, workability and efficiency of this method in apple cultivar identification.
Discussion
Although molecular markers have been used to study the genetic relationships in a variety and organisms including apple cultivars where RAPD (Palombi et al., 2002; Roche et al., 1997; Silfverberg-Dilworth et al., 2006; Stark-Urnau et al., 2002a), SSR (Palombi et al., 2002), and AFLP (Roche et al., 1997) were used, a few efficient approaches have been developed to employ DNA markers easily and efficiently in plant cultivar identification except where phylogenetic tree clusters or some fingerprints were utilized. Apparently the main reasons for these weaknesses can be attributed to the fact that no analysis could connect the information of DNA fingerprints with cultivars in an easy, clear, and readable way. We utilized the method to efficiently record DNA molecular fingerprints of genotyped plant individuals, which could be used as efficient reference information for quick plant identification. We could separate all cultivars easily just by a few corresponding primers marked on the CID.
The strategy developed and employed in the apple cultivar identification might be vital in the utilization of DNA marker in other plants as well as the development of the apple industry.
This approach has been tested and proven to be a quite efficient way of distinguishing plant cultivars and gives referable information. It is not just a simple theoretical model but a truly handy one as it makes DNA markers much more applicable for practical plant variety identification. At present, we have initiated similar work to present all the identification information of most fruit crop cultivars grown in China for use in aspects of cultivar-right-protection and for better service in the nursery and general fruit industry. The polymorphic bands generated during creation of a CID may be developed into special molecular markers for identification of the cultivar in the future. We believe that as research progresses, we
can use this method to draw a table for each species, whether plant, animal or any other organism, a table which can work just like the periodic table in chemistry but in this case providing us with the information needed to separate the species/cultivars as desired. It was possible to demonstrate that a standard set of primers can be used to distinguish many Malus domestica.
Literature Cited
Archak, S., A.B. Gaikwad, D. Gautam and E.V.V.B. Rao. K.R.M.
Swami, and J.L. Karihaloo. 2003. DNA fingerprinting of Indian cashew (Anacardium occidentale L.) varieties using RAPD and ISSR techniques. Euphytica 130:397-404.
Belaj, A., Z. Satovic, H. Ismaili, D. Panajoti, L. Rallo, and I.
Trujillo. 2003. RAPD genetic diversity of Albanian olive germplasm and its relationships with other Mediterranean countries. Euphytica 130:387-395.
Benjak, A., S. Ercisli, A. Vokurka, E. Maletic, and I. Pejic. 2005.
Genetic relationships among grapevine cultivars native to Croatia, Greece, and Turkey. Vitis 44:73-77.
Bousquet, J., L. Simon, and M. Lalonde. 1990. DNA amplification from vegetative and sexual tissues of tree using polymerase chain reaction. Can. J. For. Res. 20:254-257.
Cheng, Z.P. and H.W. Huang. 2009. SSR fingerprinting Chinese peach cultivars and landraces (Prunus persica) and analysis of their genetic relationships. Scientia Hort. 120:188-193.
Demirsoy, L., T. Demir, H. Demirsoy, Y.A. Kacar, and A. Okumus.
2008. Identification of some sweet cherry cultivars grown in Amasya by RAPD markers. Acta Hort. 795:147-152.
Ding, X.D., L.X. Lu., X.J. Chen, and X. Guan. 2000. Identifying litchi cultivars and evaluating their genetic relationships by RAPD markers. J. Trop. Subtrop. Bot. 8:49-54.
D'Onofrio, C., G. Lorenzis, T. de Giordani, L. Natali, G. Scalabrelli, and A. Cavallini. 2009. Retrotransposon-based molecular markers in grapevine species and cultivars identification and phylogenetic analysis. Acta Hort. 827:45-52.
Elidemir, A.Y. and I.U Zun. 2009. Assessment of genetic diversity of some important grape cultivars, rootstocks, and wild grapes in Turkey using RAPD markers. Acta Hort. 827:275-278.
Lee, G.P., C.H. Lee, and C.S. Kim. 2004. Molecular markers derived from RAPD, SCAR, and the conserved 18S rDNA sequences for classification and identification in Pyrus pyrifolia and P. communis. Theor. Appl. Genet. 108:1487-1491.
Lee, Y.P., G-H.Yu, Y.S. Seo, S.E. Han, Y.-O. Choi, D. Kim, I.-G. Mok, W.T. Kim, and S.-K. Sung. 2007. Microarray analysis of apple gene expression engaged in early fruit development. Plant Cell Rep. 26:917-926.
Li, R.H., B.Yang, C.S. Qiao, and B.H. Cheng. 1980. The apple varieties of Liaoning. People’s Publishing House, Liaoning, China.
Marinello, L., M.G. Sommella, A. Sorrentina, M. Forlani, and R. Porto. 2002. Identification of Prunus armeniaca cultivars by RAPD and SCAR markers. Biotechnol Lett. 24:749-755.
Melgarejo, P., J.J. Martcnez, F. HerncLndez, R. Martcnez, P.
Legua, R. Oncina, and A. Martcnez-Murcia. 2009. Cultivar
identification using 18S-28S rDNA intergenic spacer-RFLP in pomegranate (Punica granatum L.). Scientia Hort. 120:500-503.
Murray, G.C. and W.F. Thompson. 1980. Rapid isolation of high molecular weight DNA. Nucl. Acid Res. 8:4321-4325.
Palombi, M.A. and C. Damiano. 2002. Comparison between RAPDs and SSRs molecular markers in detecting genetic variation in kiwifruit [Actinidia deliciosa A. Chev]. Plant Cell Rep. 20:1061-1066.
Papp, N., B. Szilvassy, L. Abranko, T. Szabo, P. Pfeiffer, Z.
Szabo, J. Nyeki, S. Ercisli, E. Stefanovits-Banyai, and A.
Hegedus. 2010. Main quality attributes and antioxidants in Hungarian sour cherries: identification of genotypes with enhanced functional properties. Intl. J. Food Sci. Technol.
45:395-402.
Roche, P., F.H. Alston, C. Maliepaard, K.M. Evans, R. Vrielink, F. Dunemann, T. Markussen, S. Tartarini, L.M. Brown, C.
Ryder, and G.J. King. 1997. RFLP and RAPD markers linked to the rosy leaf curling aphid resistance gene (Sd1) in apple.
Theor. Appl. Genet. 94:528-533.
Sadder, M.T. and A.F. Ateyyeh. 2006. Molecular assessment of polymorphism among local Jordanian genotypes the common fig (Ficus carica L.). Scientia Hort. 107:341-351.
Silfverberg-Dilworth, E., C. Matasci, W. Van de Weg, M. Van Kaauwen, M. Walser, L. Kodde, V. Soglio, L. Gianfranceschi, C. Durel, F. Costa, T. Yamamoto, B. Koller, C. Gessler, and A. Patocchi. 2006. Microsatellite markers spanning the apple (Malus × domestica Borkh.) genome. Tree Genet. Genomes
2:202-224.
Stark-Urnau, M. 2002a. Use of RAPD-markers in Malus × domestica (apple) and Pyrus communis (pear) for cultivar identification - Part I Malus × domestica (apple). Erwerb- sobstbau 44:139-144.
Stark-Urnau, M. 2002b. Use of RAPD-markers in Malus × domestica (apple) and Pyrus communis (pear) for cultivar identification - Part II Pyrus communis (Birne). Erwerb- sobstbau 44:167-171.
Vijayan, K. 2004. Genetic relationships of Japanese and Indian mulberry (Morus spp.) genotypes revealed by DNA fingerprinting.
Plant System. Evol. 243:221-232.
Williams, J.G.K., A.R. Kubelik, K.J. Livak, A. Rafalski, and S.V. Tingey. 1990. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res.
18:6531-6535.
Xin, P.G. and F.G. Xiang. 1998. Classification of and relation for the native, introduced and bred apple cultivars in China.
J. Shandong Agri. Univ. 29:189-200.
Yonemoto, Y., A.K. Chowdhury, H. Kato, and M.M. Macha.
2006. Cultivars identification and their genetic relationships in Dimocarpus longan subspecies based on RAPD markers.
Scientia Hort. 109:147-152.
Yu, H.P., J.G. Fang, M.Y. Zhang, G. Yang, X. Cao, and H.H.
Tan. 2009. Study on application of RAPD marker in cultivar identification of seven fruit crops. Acta Agriculturae Jiangxi 21(10):5-9.