의
의
의학
학
학 박
박
박사
사
사학
학
학위
위
위 논
논
논문
문
문
P
P
Pr
r
ro
o
om
m
mo
o
ot
t
te
e
er
r
rP
P
Po
o
ol
l
l
y
y
ym
m
mo
o
or
r
rp
p
ph
h
hi
i
i
s
s
sm
m
ms
s
so
o
of
f
fC
C
Cy
y
ys
s
st
t
te
e
ei
i
i
n
n
ny
y
yl
l
l
L
L
Le
e
eu
u
uk
k
ko
o
ot
t
tr
r
ri
i
i
e
e
en
n
ne
e
eR
R
Re
e
ec
c
ce
e
ep
p
pt
t
to
o
or
r
r1
1
1(
(
(C
C
Cy
y
ys
s
sL
L
LT
T
TR
R
R1
1
1)
)
)
I
I
In
n
nf
f
fl
l
l
u
u
ue
e
en
n
nc
c
ce
e
et
t
th
h
he
e
eE
E
Ef
f
ff
f
fe
e
ec
c
ct
t
to
o
of
f
fL
L
Le
e
eu
u
uk
ko
k
o
ot
t
tr
r
ri
i
i
e
e
en
n
ne
e
e
M
M
Mo
o
od
d
di
i
i
f
f
fi
i
i
e
e
er
r
ro
o
on
n
nT
T
Tr
r
ra
a
an
n
ns
s
sc
c
cr
r
ri
i
i
p
p
pt
t
ti
i
i
o
on
o
n
na
a
al
l
lR
R
Re
e
eg
g
gu
u
ul
l
l
a
a
at
t
ti
i
i
o
o
on
n
n
o
o
of
f
ft
t
th
h
he
e
eC
C
Cy
y
ys
s
sL
L
LT
T
TR
R
R1
1
1G
G
Ge
e
en
n
ne
e
eI
I
In
n
nd
d
du
u
uc
c
ce
e
ed
d
db
b
by
y
yI
I
IL
L
L-
-
-4
4
4
아
아
아 주
주
주 대
대
대 학
학
학 교
교
교 대
대
대 학
학
학 원
원
원
의
의
의 학
학
학 과
과
과
오
오
오 정
정
정 미
미
미
오
오
오정
정
정미
미
미의
의
의 의
의
의학
학
학 박
박
박사
사
사학
학
학위
위
위 논
논
논문
문
문을
을
을 인
인
인준
준
준함
함
함.
.
.
심
심
심사
사
사위
위
위원
원
원장
장
장
강
강
강
엽
엽
엽
인
인
인
심
심
심 사
사
사 위
위
위 원
원
원
박
박
박 해
해
해 심
심
심
인
인
인
심
심
심 사
사
사 위
위
위 원
원
원
남
남
남 동
동
동 호
호
호
인
인
인
심
심
심 사
사
사 위
위
위 원
원
원
김
김
김 승
승
승 현
현
현
인
인
인
심
심
심 사
사
사 위
위
위 원
원
원
지
지
지 영
영
영 구
구
구
인
인
인
아
아
아 주
주
주 대
대
대 학
학
학 교
교
교 대
대
대 학
학
학 원
원
원
2
2
20
0
00
0
06
6
6년
년
년 6
6
6월
월
월 2
2
22
2
2일
일
일
Promoter Polymorphisms of Cysteinyl Leukotriene Receptor
1 (CysLTR1) Influence the Effect of Leukotriene Modifier on
Transcriptional Regulation of the CysLTR1 Gene Induced by
IL-4
by Jung-Mi Oh
A Dissertation Submitted to The Graduate School of Ajou University
in Partial Fulfillment of the Requirements for the Degree of
DOCTOR OF PHILOSOPHY
Supervised by
Hae Sim Park, M.D., Ph.D.
Department of Medical Sciences
The Graduate School, Ajou University
- ABSTRCT -
Promoter Polymorphisms of Cysteinyl Leukotriene Receptor 1
(CysLTR1) Influence the Effect of Leukotriene Modifier on
Transcriptional Regulation of the CysLTR1 Gene Induced by IL-4
Background: In recent, intensive studies have attempted to elucidate the genetic
background of aspirin intolerant asthma (AIA), but genetic factors associated with its pathogenesis are still not completely understood. Cysteinyl leukotrienes (CysLTs) play important roles in eosinophilic airway inflammation of AIA via CysLTR1-mediated mechanism. Leukotriene modifier MK-571, a selective CysLTR1 antagonist, is widely used in the management of asthma. We selected Cysteinyl leukotruene receptor 1 (CysLTR1) among leukotriene – related genes and screened for genetic variation in the 5’ upstream region of CysLTR1 gene. This study was designed to investigate whether MK-571 possesses inhibitory effect on transcriptional induction of CysLTR1 induced by IL-4 or not.
Materials and methods: Genetic variation in 5’ upstream region of CysLTR1 gene
were screened in 43 normal healthy controls by direct sequencing and then genetic association study of CysLTR1 genetic polymorphisms were performed in 105 aspirin intolerant asthma (AIA) patients, 111 aspirin tolerant asthma (ATA) patients, and 125 normal healthy controls (NC) . Promoter activities according to the genetic polymorphism were examined by luciferase reporter assay. Transcriptional
regulation of endogeneous CysLTR1 gene induced by IL-4 was investigated using lung carcinoma cell line, A549 cells, by real time PCR and flowcytometry. Effect of MK-571 on the transcriptional induction of CysLTR1 by IL-4 was examined by luciferase reporter assay.
Results: In the screening of 5’ upstream region of CysLTR1 gene in 43 normal
healthy controls, three single nucleotide polymorphisms (SNPs) including C-634T, A-475C, and A-336G were identified. Though a genetic association studies of 3SNPs in 105 aspirin -intolerant asthma (AIA) patients, 111 aspirin tolerant asthma (ATA) patients, and 125 normal healthy controls (NCs), significant association of 3 SNPs with AIA were found. There were significant differences in allele frequencies of the 3 SNPs within male subjects; the common 3 SNP haplotypes, ht1 [C-A-A], was associated with decreased disease risk (p=0.031 for AIA vs. NC; p=0.022 for AIA vs. ATA), and the ht2 [T-C-G] haplotype with increased disease risk (p=0.031 for AIA vs. NC; p=0.031 for AIA vs. ATA). Moreover, 3 SNPs can modulate CysLTR1 gene expression; promoter activity was enhanced with ht2 [T-C-G] construct in comparison of ht1 [C-A-A] construct in jurkat cells, U937 cells, and A549 cells. Endogeneous level of CysLTR1 gene was remarkably increased by IL-4 in A549 cells. Promoter activity was also significantly increased with the ht2 [T-C-G] construct in comparison with ht1[C-A-A] construct in IL-4 primed A549 cells (p=0.008), which was significantly inhibited by pre-treatment with MK-571(p=0.004), not with dexamethasone in which ht1 [C-A-A] construct promoter activity was not significantly inhibited by dexamethasone.
Conclusion: These results suggest that genetic variants of CysLTR1, ht2 [T-C-G],
can up-regulate CysLTR1 expression and significantly induced CysLTR1 expression by IL-4. MK-571 may be more beneficial to control asthmatic symptoms, especially in patients carrying variant genotype of the CysLTR1 promoter.
TABLE OF CONTENTS
ABSTRACT ... ⅰ
TABLE OF CONTENTS ... iv
LIST OF FIGURES ... vi
LIST OF TABLES ... vii
. Ⅰ INTRODUCTION ... 1
. Ⅱ MATERIAL AND METHODS ... 8
A. Subjects ... 8
B. Identification of genetic polymorphism in CysLTR1 gene ... 8
C. Plasmid construction for promoter activity and transfection ... 10
D. Nuclear extracts preparation and electrophoretic mobility shift assay ... 11
1. Nuclear extracts preparation ... 11
2. Electrophretic mobility shift assay (EMSA) ... 12
3. Sequence mutation analysis ... 13
E. CysLTR1 promoter deletion and luciferase reporter assay ... 14
F. Phamacological treatments and reporter gene assay... 15
G. CysLTR1 expression by RT-PCR, Realtime-PCR and flowcytometry ... 16
1. RT-PCR analysis for CysLTR1 mRNA ... 17
2. Realtime-PCR analysis for CysLTR1 mRNA ... 17
3. CysLTR1 expression by flowcytometry ... 18
H. Statistical analysis ... 19
. Ⅲ RESULTS ... 20
A. Identification of three genetic polymorphisms in CysLTR1 gene ... 20
B. Genetic association study of CysLTR1 genetic polymorphism ... 20
1. Clinical characteristics of the study subjects ... 20
2. Genotype distribution of CysLTR1 3 SNPs ... 23
3. Association analysis of AIA associated quantitative phenotypes ... 25
4. Effect of 3 SNPs on transcriptional activity ... 28
5. Identification of unknown nuclear protein binding by EMSA ... 28
C. Characterization of CysLTR1 promoter in A549 cells... 31
D. Transcriptional regulation of CysLTR1 gene by IL-4 ... 33
1. Expression of CysLTR1 ... 33
2. Effect of genetic polymorphism on the transcription regulation by IL-4 33 E. Effect of leukotriene modifier on the transcription regulation by IL-4 ... 36
F. Effect of dexamethasone on CysLTR1 mRNA expression in A549 cells ... 38
. Ⅳ DISCUSSION ... 41 . Ⅴ CONCLUSION ... 49 REFERENCE ... 50 국문요약 ... 64
LIST OF FIGURE
Fig. 1. AIA pathogenesis mechanism, COX theory ... 2 Fig. 2. 5’-RACE sequence and 3 SNPs location on the 2466bp fragment of CysLTR1gene. ... 21 Fig. 3. Gene map and linkage disequilibrium (LD) coefficients in cysteinyl leukotriene receptor 1 (CysLTR1) ... 22 Fig. 4. Expression of CysLTR1 mRNA in various cell lines ... 29 Fig. 5. Promoter activity assay of human CysLTR1 promoter constructs ... 30 Fig. 6. Unknown factor bind to a -475A, -336A, and -336G region in the CysLTR1
promoter ... 32 Fig. 7. Deletion analysis of the human CysLTR1 promoter ... 34 Fig. 8. CysLTR1 mRNA and protein expression ... 35 Fig. 9. Drug effect by dexamethasone and MK-571 on CysLTR1 promoter activity ... 37 Fig. 10. Inhibitory effect of MK-571 on IL-4 induced CysLTR1 luciferase activity in A549 cells. ... 39 Fig. 11. Realtime PCR analysis of effects on CysLTR1 mRNA with addition of dexamethasone in A549 cells ... 40
LIST OF TABLES
Table 1. Clinical characteristics of the study subjects ... 24 Table 2. Genotype frequencies of the three SNPs in CysLTR1 promoter ... 26 Table 3. Association of CysLTR1 C-634T with atopy, total IgE, asthma duration,
I. INTRODUCTION
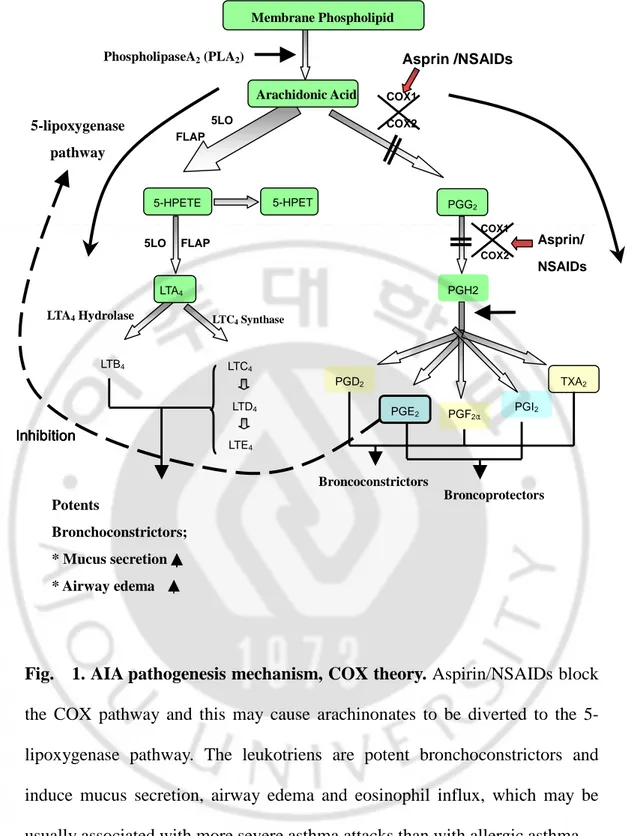
In a subset of asthmatic patients, aspirin and several other nonsteriodial anti-inflammatory drugs (NSAIDs) that inhibit cyclooxygenase enzymes (COXs) induce severe asthmatic attacks, generally termed aspirin -intolerant asthma (AIA) (Szczeklik and Stevenson,1999). AIA is characterized by distinct clinical syndrome ‘aspirin triads’, aspirin -hypersensitivity, bronchial asthma, and chronic rhinisinusitis with nasal polyposis, which is usually more severe asthma attacks, than in allergic asthma. AIA is seen in approximately 5-15% of adult asthmatic patients in commonly begins in middle age and more prevalent in women (Sampson and Szczeklik, 1998, Picado, 2002; Szczeklik and Kielbasa, 2004). Although the mechanism of aspirin sensitivity in asthma remains unclear, most investigator are widely accepted that aspirin is involved in the development of asthma attacks in patients with AIA not through allergic mechanism but through a pharmacological mechanism, because drug that inhibits cyclooxygenase-1 (COX-1), but not COX-2, induced bronchoconstriction in patients with AIA and there is no evidence of an IgE response to aspirin (Cowburn et al., 1998; Lam and Frank Austen, 2000; Pierzchalska and Szczeklik 2003). The pathogenic mechanism of AIA has been explained by cyclooxygenase theory (Fig. 1.): Pharmacological actions of NSAIDs, inhibition of COXs in respiratory tracks, alter arachidonic acids (AA) methabilosm in AIA patients. Aspirin and most NSAIDs lead to decrease an anti-inflammatory effect of PGE2 (Sestini et al., 1996; Celik and Melli, 2001), finally leading to increase production of Cysteinyl leukotrienes (CysLTs)
Arachidonic Acid 5-HPETE 5-HPET LTA4 Membrane Phospholipid PhospholipaseA2 (PLA2) FLAP LTB4 LTC4 LTD4 LTE4 PGG2 5LO 5LO FLAP
LTA4 Hydrolase LTC4 Synthase
Asprin /NSAIDs COX1 COX2 PGH2 PGD2 PGE2 PGF2α PGI2 TXA2 5-lipoxygenase pathway COX1 COX2 Asprin/ NSAIDs Broncoconstrictors Broncoprotectors Inhibition Inhibition Inhibition Inhibition Potents Bronchoconstrictors; * Mucus secretion * Airway edema * eosinophil influx
Fig. 1. AIA pathogenesis mechanism, COX theory. Aspirin/NSAIDs block
the COX pathway and this may cause arachinonates to be diverted to the 5-lipoxygenase pathway. The leukotriens are potent bronchoconstrictors and induce mucus secretion, airway edema and eosinophil influx, which may be usually associated with more severe asthma attacks than with allergic asthma.
that can mediates bronchoconstrcition, mucus secretion, vascular permeability, cellular infiltration, and eosinophil survival (Biagaard H, 2000; Szczeklik and Kielbassa, 2004). These studies suggest that AIA is most likely to be related to abnormalities of prostanoid and eicosanid methabilosms, and overproduction of CysLTs and overexpression of the CysLT receptors on inflammatory cells were noted within the respiratory tracks (Sousa and Lee, 2002).
Cysteinyl leukotrienes (CysLTs, LTC4, LTD4, and LTE4) belong to a large family of lipid mediator (Samuelsson, 1983; Drazen and Austen, 1987), termed eicosanoids, that are arachidonic acid (AA) released from the cell membrane by phospholipases. The oxygenation of AA to form 5-hydroperoxyeicosatretranoic acids and the immediate dehydration of this unstable intermediate to LTA4, is carried out
by 5-lipoxygenease (5-LO). LTA4 produced from AA by 5-LO is the common
substrate from which all CysLTs are formed. Metabolism of LTA4 by LTA4 hydrolase
(LTA4-H) result on the production of the potent chemoattractant LTB4. Alternatively,
LTA4 can be conjugated with glutathione by LTC4 synthase (LTC4S) to produce LTC4, LTD4 and LTE4. These lipid mediators were found at high level in most inflammatory cells, including neutrophils, monocytes, macrophages, mast cells, basophils, and eosinophils in the process of allergic inflammation. CysLTs stimulate mucus secretion, induce bronchial hyperresponsiveness, cause constriction of airway smooth muscle, eosinophils recruitment into the airways, and increase vascular permeability (Dahlen and Samuelsson, 1980; Piper, 1984; Lewis and Soberman, 1990; Piper and Price, 1991), which was participated in the pathogenesis of
inflammatory diseases, including bronchial asthma, arthritis, and rhinitis (Samuelsson, 1983; Drazen and Austin, 1987; Hay et al., 1995; Horwitz et al., 1998).
The CysLTs exert their effects through cell surface receptors, which were classified into two major subtypes: Cysteinyl leukotriene receptor 1 (CysLTR1; MIM300201; GenBank accession number, AF 119711; Lynch et al., 1999) and Cysteinyl leikotriene receptor 2 (CysLTR2), and they have been cloned (Coleman et al., 1995; Metters, 1995). Both receptors belong to the rhodopsin family of the G-protein coupled receptor (GPCR) gene superfamily (Crooke et al., 1990; Metters, 1995), these bind to LTC4, LTD4, and LTE4 with affinity order LTD4>>LTC4>LTE4 (CysLTR1) and LTC4 = LTD4>>LTE4 (CysLTR2), both are expressed on bronchial smooth muscle cells and inflammatory leukocytes including peripheral blood monocytes and eosinophils (Del Prete, et al. 1988; Schleimer et al.,1992; Chibana and Fukuda, 2003). CysLTR1 gene is located on chromosome Xq13-q21 (Lynch et al., 1999), and the cDNA encodes 1014bps with a calculated molecular mass of 38,549 Da. Responses are produced by simulation of CysLTR1, which is sensitive to leukotriene modifiers such as montelukast (MK-476; Singulair) (Jones et al., 1995; Reiss et al., 1996), zafirlukast (ICI 204,219; Accolate) (Krell et al., 1990; Spector et al., 1994), and pranlukast (Onon; SB 205312; Ono-1078) (Obata et al., 1992; Tamaoki et al., 1997), pobilukast ( Hay DW et al., 1987), and MK-71 (Jones TR et al. ,1987) and these leukotriene modifiers cannot antagonize responses through CysLTR2. There have been reports that the overproduction of CysLTs has been demonstrated in the airways of
AIA patients (Smith CM and Lee TH, 1992), The enhanced activities of key synthetic enzymes, such as 5-LO and LTC4S (Sampson, 1997), have been implicated in the elevated production of CysLTs in patients with AIA. For example, LTC4S, the terminal enzyme in the production of CysLTs, is markedly over-expressed in eosinophils and mast cell in airway mucosa of patients with AIA (Szczeklik and Stevenson, 1999). Total number of inflammatory leukocytes expressing CysLTR1 was significantly elevated in nasal mucosa of patients with aspirin-intolerant chronic rhino sinusitis compared with aspirin-tolerant chronic rhino sinusitis (Sousa and Lee, 2002). The topical nasal desensitization of the AIA associated with a reduction in the number of nasal inflammatory cells expressing CysLTR1 (Steinke et al., 2003), suggesting that over expression of CysLTR1 was probably related with pathogenesis of aspirin hypersensitivity. CysLTR1 selective antagonist could block bronchoconstriction induced by lysine aspirin in some patients with AIA (Dahlen B et al, 1993). These findings suggest that overproduction of CysLTs and CysLT receptors may play roles in the pathogenic mechanism of AIA, which are inhibited proinflammatory effect by selective CysLTR1 antagonists, which have been considered as key drugs in the management of AIA.
In addition, CysLTs promoted generation of Th2 cytokines such as IL-4, IL-5, IL-13 and those cytokines also enhanced generation of CysLTs in macrophages, eosinophils, and monocytes by inducing the expression of CysLTR1 (Thivierge and Rola-Pleszczynski, 2000; Thivierge and Rola-Pleszczynski, 2001; Chibana and
Fukuda, 2003; Peters-Golden and Sampson, 2003). IL-4 may also promote tissue
eosinophila by vascular cell adhesion molecule-1 (VCAM-1; Schleimer et al,
1992), is an essential cofactor for IgE switching in B lymphocyte (Del Prete, et al, 1988). There were extensive interactions between CysLTs and other mediators
such as macrophage inflammatory protein (MIP)-1β (Frieri et al., 1999), TNF-α
(Elizabeth and Joshu, 2002), NF-κB, and RANTES (Kawano T et al., 2003), these
are relevant to asthmatic inflammation via a CysLTR1–mediated mechanism. These previous reports demonstrated that expression of CysLTR1 mRNA can be increased by those Th2 inflammatory cytokines, implying a positive feedback by endogenously produced CysLTs. That is, in a Th2 cytokine -rich environment, CysLTR1 expression can be up-regulated which promotes proinflammatory effects of CysLTs. These findings suggested that CysLTR1 might be one of the key genes in pathogenic mechanism of AIA. Although CysLTR1 gene was suggested to have a major role in pathogenesis of AIA, there has been no previous analysis of association between CysLTR1 polymorphism and aspirin intolerance in asthma. In an effort to discover polymorphism(s) in CysLTR1 gene, we screened for genetic variation in the 5’ upstream region of CysLTR1 by direct sequencing and identified 3 novel SNPs (C-634T, A-475C, A-336G), which were located in the 5’ upstream region of the gene at positions -634, -475, -336 from the transcription start site of CysLTR1. Next, to investigate whether these genetic variants of the CysLTR1 gene are related to the AIA phenotype, we conducted a case-control association study of 3 groups including of Korean subjects classified as AIA, aspirin-tolerant asthmatics (ATA), and healthy controls (NCs). Furthermore, we
investigated the response of the CysLTR1 promoter gene induced by Th2 cytokine, IL-4, and effect of MK-571 on transcriptional induction of CysLTR1 by IL-4.
I. MATERIALS AND METHODS
A. Subjects
Three groups of the study patients (105 patients with AIA, 111 patients with ATA and 125 normal healthy controls) were enrolled from the department of Allergy and Rheumatology, Ajou University Hospital in Korea. All subjects were of Korean ethnicity. Diagnosis of asthma was confirmed using the Global Initiative for Asthma (GINA) guidelines (2004, revised) and diagnosis of AIA was based on those showing positive response to lysine aspirin (L-ASA) bronchoprovocation tests, which was performed according to a modified method as previously described (Choi JH et al., 2004). All subjects gave informed consent and the protocol was approved by the local ethics committee. Skin prick tests were performed using 12 common aeroallergens (Bencard, UK), as well as positive (histamine) and negative (saline) controls. Atopy was defined as one or more positive reactions on skin prick test result. All the clinical histories were reviewed in detail by the investigators. Methacholine bronchial challenge tests were performed according to a previously described method (Choi JH et al., 2004).
B. Identification of genetic polymorphism in CysLTR1 gene
Genetic variation in 5’ upstream region of CysLTR1 gene were screened in 43 normal healthy controls by direct sequencing, this results we found 3 SNPs in CysLTR1 gene. The transcription start site of the human CysLTR1 transcript was
CA, USA). Nucleotide position on (+1) is transcription start site determined by 5’-RACE. In the end, we identified three polymorphic sites: C-634T, 475C, A-336G from the transcriptional start site.
In brief, for the 5' rapid amplification of cDNA ends (RACE), human fetus marathon ready cDNA (Clontech, Palo Alto, CA, USA) was ligated to an adaptor
cassette at 5' ends with the SmartTM RACE cDNA Amplification Kit (Clontech,
Palo Alto CA, USA). The 5' RACE was performed by PCR with an adaptor primer (Clontech, Palo Alto CA, USA) and human CysLTR1 gene-specific primers: sense primer, AP1; antisense primer, 5'AAGTAGATCTGC TACTGC3'and 5' -TCATCAATAG TGTCA TGGC-3'. The PCR products were diluted (1:50) to performed secondary PCR with the primer pairs: sense primer AP-2; antisense primer, 5'-TCGATCTA CCAGTCCTTG C -3' and 5'- ATTCACAGAGT GCCTGCTC-3'. PCR product was cloned into the Topo II dual promoter TA cloning vector (Invitrogen, Groningen, Netherlands). Positive cleavage samples were sequenced using Big Dye Terminator sequencing (ABI 377 sequencer; Applied Biosystems, Foster City, CA, USA).
Genomic DNA was extracted from peripheral leukocytes isolated from acid citrate dextrose solution (ACD) anti-coagulated blood through use of a
Ficoll-PaqueTM PLUS (Amersham Biosciences, Uppsala, Sweden) gradient method.
Peripheral blood (10ml) was collected from the participant of this study. The leukocytes were layered on top of two discontinuous gradients of Ficoll PaqueTM PLUS and the tube was centrifuged at 2600 rpm for 20 min at 25℃. After the cells
in the upper layer, this consisted of lymphocytes, monocytes and platelets were collected and washed with 1X phosphate-buffered saline (PBS) twice. Genomic
DNA was used in G-DEXTM for Blood Genomic DNA Extraction Kit (iNtRON
Biotechnology, Korea) according to the manufacturer's instructions. 5’-untranslated region of the CysLTR1 gene was amplified by the primers 5'- CATTTGGGAATGGGTGAATC-3' and 5'-CTGCTAACTTCAAGGTCCA-3’. PCR was performed, in a 50ul reaction mixture containing 100ng of genomic DNA,
2.5mM MgCl2 0.2uM each primer, 0.2mM dNTPs, 2mM betaine (Sigma-Aldrich,
USA), 5% DMSO and 0.5 U LA-Taq (Takara Shuzo, Japan) under the following conditions; denaturation at 95℃ for 10 seconds, annealing at 60℃ for 1 minute, extension at 72℃ for 3 minutes, for 50 cycles. A 2466bp fragment of the CysLTR1 (corresponding to - 2158 to +313) was scanned for the presence of polymorphisms by direct sequencing within three groups of individuals classified as aspirin-intolerant, aspirin-tolerant asthmatic, and control healthy individuals. In the end, we identified three polymorphic sites; C-634T, A-475C, A-336G from the transcriptional starting site.
C. Plasmid construction for promoter activity and transfection
A 2466bp fragment (from -2158 to +313) of the human CysLTR1 gene promoter was prepared by PCR using two haplotype forms (ht1 [C-A-A] and ht2 [T-C-G]; C-634T, A-475C, A-336G) of genomic DNA as a template, each of which was separately subcloned into an KpnI-XhoI site of the pGL3-Basic luciferase reporter vector (Promega Corp., Madison, WI, USA). A transfection experiment into Jurkat
cells (human T lymphocytes, No. 40152, KCLB, Korea) was undertaken nine times (27 in total) , A549 (human lung carcinoma cell, No. 10185, KCLB, Korea) (30 in total), and U937 cells (Human monocytes, 12 in total) .
Next, to performed luciferase activity, Jurkat cells, U937, and A549 cells were cultured in RPMI 1640 with 10% FBS (Gibco-BRL, Paisley, UK), 100units/ml penicillin, and 100ug/ml streptomycin at 37’C in a humidified atmosphere with 5% CO2. The cells were grown in growth medium and seeded in six-well plates. At 24h after seeding, cells were harvested and washed with OPTI-MEMI reduced serum medium (Life Technology, USA). In each transfection, 2.5µg of the pGL3 constructs 1µg of pSV beta-galactosidase expression vector (Promega, Madison, WI) and 10ul lipofectamin (Life Technology, USA) were used. At 48h after transfection, cells were lyses and assayed for the firefly luciferase activity using luciferase assay system (Promega, Madison, WI) by according to the manufacturer’s instruction. Luciferase activity was normalized to beta-galactosidase activity, and the relative luciferase activity of the reporter constructs was expressed in relation to the activity of the pGL3-basic vector. Data are the mean values of three independent experiments, plus or minus the SD.
D. Nuclear extracts preparation and electrophoresis gel mobility shift assay
1. Nuclear extracts preparation
Nuclear extracts were prepared from THP-1 and U937cells that were untreated and treated with mitogens phorbol 12-myristate 13-acetate (PMA) and
10ng/ml IL-4 (Sigma Aldrich), as described by Schreiber et al (Schreiber and
Schaffner, 1989) with some modification. Briefly, 5×106 cells were resuspended in
400 µl of cold hypo-osmotic buffer [10 mM Hepes /KOH, 2 mM MgCl2, 0.1mM
EDTA, 10 mM KCl, 1 mM DTT (dithiothreitol), 1 mM PMSF and protease inhibitors (Roche Molecular Biochemicals), pH 7.9] and left on ice for 10 min. Then, Nonidet P40 (0.007%) was added to the cell suspension that was mixed by vortex and centrifuged at 15000g for 30 sec. The supernatant containing the cytoplasmic proteins could be stored at −80℃. The pellets of nuclei were gently
resuspended in 15 µl of cold saline buffer [50 mM Hepes/KOH, 50 mM KCl, 300
mM NaCl, 0.1 mM EDTA, 10% (v/v) glycerol, 1 mM DTT, 1 mM PMSF and protease inhibitors (Roche Molecular Biochemicals), pH 7.9] and left for 20 min on ice. After centrifugation at 15000g for 15 min, at 4℃, the supernatant containing nuclear proteins was stored at −80℃. Protein concentrations were measured with the Bradford protein assay kit from Bio-Rad.
2. Electrophretic mobility shift assay (EMSA)
To investigate whether the SNPs created nuclear transcription protein, we
carried out EMSA. In brief, 5 µg of nuclear proteins was incubated for 30 min at
room temperature (25℃) in a volume of 10 µl with 0.2 ng of labelled
oligonucleotide probe in binding buffer [20 mM Hepes/ KOH (pH 7.9), 75 mM
NaCl, 1 mM EDTA, 5%glycerol, 0.5 mM MgCl2 and 1 mM DTT] containing 2 µg
of BSA and 400ng of poly (dI-dC) (Amersham Biosciences, Roosendael, The Netherlands). DNA–protein complexes were then resolved by electrophoresis on a
non-denaturing 6% (w/v) polyacrylamide gel for 2 h at 300 V in 0.5×TBE (2.5 mM
Tris, 2.5 mM H3BO3 and 2 mM EDTA). The gels were then dried and auto-radio
graphed on an IP-plate (Fuji photofilm Co., Ltd., Tokyo).The image produced by the FLA3000 scanner was analyzed using image-gauge ver.4 software (Fuji photofilm Co., Ltd., Tokyo). The sequences of the oligonucleotide probes were as follows: CysLTR1 C634T probe, 5’CTGCTATG TCCC[c/t]TTTCTCCAGAAG -3’; CysLTR1 A-475C probe, 5’-GAAA[a/c]AACAGTTTTAATCCAATGG-3’;CysLTR1 A-336G probe, 5’-AGAGAGAGAGAGA
AAC[a/g]GGAGAGATG-3’, and NF-κB probe;GGTTACAAGGGACTTTCCGCTG-3 and
5’-TTGGCAGCGGAAAGTCCCTTGT-3’.To the determine the specificities of the DNA complexes, 1.75uM unlabeled probe was incubated with the nuclear extract for 20min before addition of the labeled probe. The oligonucleotide probes were labelled by infilling with the Klenow DNA polymerase ( Promega Corp., Madison, WI, USA).
3. Sequence mutation analysis
To determine whether these SNP of CysLTR1 created or losed a transcription factor binding site, sequences were submitted to the ‘Signal Scan’ online program
(www.bimas.cit.nih.gov/molbio/signal) and compared with mammalian
transcription factor binding sequences in the transcription factors (TRANSFAC) database. There was no transcription factor binding site matched on ‘Signal Scan’ online program and TRANSFAC data base.
E. CysLTR1 promoter deletion constructs and luciferase reporter assay
Region of interest were amplified from above 2466bp plasmid by PCR other primers containing consensus sequence for SacI and SmaI restriction enzymes using pfu polymerase (Solgent), under the following pairs of primers were used: B567 construct: (C1) 5’- ACGGAGCTC TCCCCACC TACAAATAGTCTAATG-3’/(R1) CGGCCCG GGCGCTGCTAACTTCAAGGTCCA-3’; B981:(C2) 5’-ACGGAGCTCCACATACCAGACTGCTATG-3’/(R1); B1570: (C3) 5’- ACGG AGCTCACTGTAGCTCAAAATGGCTG-3’/(R1); B808: (C2)/(R2) 5’- CGGCCC GGGATTCACAGAGTGCCTGCTC-3’. PCR products were gel purified with agarose gel purification kit (iNtRON, Korea), cut with the restriction enzymes (Takara, Shuzo, Japan) and ligated into pGL3 basic vector using T4 DNA ligase (Elpis Biotech, Korea). All constructs were verified by sequencing. Plasmid DNAs were prepared from these construct using the Endo Free plasmid Maxi kit (Qiagen) and quantities and integrity by UV spectrometry and gel loading.
A549 cells were grown in growth medium and seeded in 12-well plates. In each transfection, 1µg of the plasmid constructs 0.5µg of pSV beta-galactosidase expression vector (Promega, Madison, WI) and 4ul lipofectamin (Life Technology) were used. At 48h after transfection, cells were lysed and assayed for the firefly luciferase activity (Promega Madison, WI) by according to the manufacturer’s instruction. Luciferase activity was normalized compared to beta-galactosidase activity, and the relative luciferase activity of the reporter constructs was expressed in relation to the activity of the pGL3-basic vector. Data are the mean values of
three independent experiments, plus or minus the SD.
F. Pharmacological treatments and reporter gene assay
A549 cells were cultured in RPMI 1640 medium supplemented with 10% heat -inactivated FBS (Gibco/ BRL), 100 U/ml penicillin, 25 mM Hepes, and 100 mg/ml streptomycin in a humidified atmosphere with 5% carbon dioxide (CO2) at 37°C. Cells were cultured overnight in complete media before changing to fresh serum free media containing IL-4 (10ng/ml) (Sigma-Aldrich, USA). MK-571 and
dexamethasone (all from Sigma-Aldrich, USA) were dissolved in
dimethylsulfoxide (DMSO) and absolute ethanol. Final concentrations of DMSO / ethanol added to cells were <0.1% and this had no effect on any of the responses (data not shown). The day before each experiment, cells were seeded into 12 well and 6 well tissue culture dishes the density of 1×105 cells. For the evaluation of dexametasone and MK-571, the cells were incubated with various concentrations of dexamethasone and MK-571 for 24hr before the stimulation by IL-4 (10ng/ml). Transient transfections were performed using the lipofectamine reagent (Invitrogen) according to the manufacturer’s suggested protocol. To correct for the different transfection efficiencies of the various luciferase construct, the pSV beta-galactosidase expression vector plasmid (Promega, Madison, WI) was co-transfected with the CysLTR1 Promoter luciferase construct into the cells. Briefly,
1×105 cells (per wells) were seeded into 12 well tissue culture plates before the day
of transfection in RPMI1640 medium plus 10% FBS. For each well 1µ g CysLTR1 promoter luciferase construct and 0.5ug pSV beta-galactosidase plasmid was
co-transfected into the cells in and 4ul lipofectamine (Gibco BRL) into the cells in 1ml serum free medium. After 5h of transfection, 1 ml medium containing 20% FBS was added. And the cells were further incubated for a total transfection time of 48h. After at 48h, the RPMI 1640 medium was replaced. Then serum free medium, stimulant, stimulant plus drugs (dexamethasne and MK-571) were added to the transfected cells. Luciferase activity was measured using the firefly luciferase reporter assay system (Promega Madison, WI). Briefly, 100ul of lysis buffer was added to each well and plates were agitated for 15min at room temperature. 20ul of supernatant was added to 100ul luciferase assay reagent and light unit measured for 10s at three times on DLR-Ready. The promoter activity was calculated as luciferase activity / beta-galactosidase activity, and expressed as fold of increase relative to the activity of the pomoterless pGL3-basic vector.
In experiments where the effect of stimulant (IL-4, 10ng/ml), dexamethasone, and MK-571(CysLTR1 specific antagonist) on luciferase activities were studied, both the control vector-transfected (PGL3 basic vector) and promoter construct-transfected cells were treated with the corresponding drug in culture medium containing 2% FBS for the designated periods time before doing the beta-galactosidase and luciferase assay. The pharmacological reagents used in this study were purchased from the Sigma-Aldrich Corp. (Oakville, Ontario, Canada). Data are presented as the mean values of three independent experiments, plus or minus the SD.
1. RT-PCR analysis for CysLTR1 mRNA
Total RNA was extracted with the use of Easy-Blue reagent (iNtRON Biotechnology, Korea) from A549 cells cultured in 6-well tissue culture plates. The 4ug of total RNA was converted to cDNA by the reverse transcribe using the MMLV-reverse transcriptase (Promega, Madison, WI) according to the manufacturer’s suggested protocol. After incubation at 42°C for 90min, the samples were heated for 10min at 72°C to terminate the reactions and were stored at -20°C until used. PCR was performed with DNA Engine thermal cycler (MJ research, Inc., Biozym) for 32 cycles consisting of 2 min denaturation at 94°C, 30 s annealing at 55°C, and 30 s extensions at 72°C, followed by a final 10min extension at 72°C. CysLTR1 was amplified with the primers derived from the cDNA sequence for CysLTR1: 5’-CTTTGTGCTCTAT GTCCTCA-3’ as sense and 5’ CACGACCATGATCATTCCTA-3’ as antisense. Samples were subjected
to parallel amplification of the constitutively expressed, housekeeping gene,
β-actin using the following primers: 5’- TCCTTCTGCATCCTGTCG GCA -3’ as sense and 5’-CAAGAGATGGCCACGGC TGCT-3’as antisense. A 10ul aliquot from each PCR was allowed to migrate by electrophoresis in a 1% agarose gel. The CysLTR1 amplified fragment contained 600 bps. The gel was then colored with ethidium bromide and photographed under UV transillumination. No PCR products were obtained when reverse transcriptase was omitted, indicating that there was no DNA contamination.
The realtime PCR were carried out on the on ABI Prism 7500 sequence detection system (Applied Biosystems), using the SYBR green dyes (Molecular probe), according to the recommendations of the vendors. The primer combination of CysLTR1 gene were as followed: 5’- TGACCGCTGCCTTTTTAGTC-3’ as sense and 5’ GAGAGGGTCAAAGCAACAATTG-3’as anti-sense, this amplified fragment contained 192bps. PCR protocol was applied: 15min hot start at 95℃, followed by 40 cycles of denaturation (30s at 94℃), annealing (30s at 60℃) and synthesis (30s at 72℃, total volume of 20ul). For melting curve analysis, the temperature was elevated slowly from 60℃ to 95℃. Data were acquired and analyzed with the ABI 7500 prism software, the amplication kinetics were recorded as sigmoid progress curves, for which the fluorescence was plotted against the number of amplication cycles. The threshold cycle number (CT) was used to define the initial amount for each template. CT was the first cycle, for which a detectable fluorescence signal was observed and relative gene expression was calculated as a fold induction compared with control.
3. CysLTR1 expression by flow cytometry
The expression of CysLTR1 in A549 cells and IL-4 primed A549 cells was assessed using a polyclonal anti-CysLTR1 antibody (Cayman) directed against the carboxyl-terminal portion of the receptor. The antibody was raised against a peptide corresponding to amino acids 318–337 of the C terminus of human CysLTR1. For flow cytometry studies, A549 cells were washed with PBS and fixed with 2% paraformaldehyde for 15 min at room temperature followed by
permeabilization with 0.1% Triton X-100 for an additional 15 min at room temperature. Cells were resuspended with PBS/2% BSA and labeled for 30 min at 4°C with anti-CysLTR1 antibody (or with control, non-pertinent Ab). Cells were then washed with cold PBS and incubated for 30 min at 4°C with FITC-conjugated goat anti-rabbit IgG (SantaCruiz). Finally, cells were washed again and resuspended in PBS before single-color immunofluorescence analysis of 10000 cells was performed on a FACS vantage (BD). A 1/1000 dilution of the anti-CysLTR1 antiserum was used in all flowcytometry studies.
H. Statistical analysis
Genotype–phenotype association between CysLTR1 SNPs and asthma-associated phenotypes was analyzed with the SPSS version 11 package (SPSS Inc., Chicago, IL, USA). The biallelic SNPs were coded into two (dominant, recessive model) or three (additive model) classes and analyzed categorically relative to the most common homozygous genotype for each SNP. Bivariate analysis used analysis of variance (ANOVA) or t-tests to compare continuous outcomes across the levels of each genotype and w2 tests and calculation of odds ratios (ORs) with 95% confidence intervals (CIs) on contingency tables when comparing genotype to categorical variables. Among control subjects, Hardy–Weinberg equilibrium was tested at each SNP locus by a w2 goodness-of-fit test. Pairwise linkage disequilibrium (LD) between SNP loci was measured using both the absolute value of Lewontin’s D0 and r2 (Hedrick PW, 1987). Haplotypes of the CysLTR1 gene were analysed using Haploview v2.0 (Barrett JC and Daly, 2005).
III. RESULTS
A. Identification of three genetic polymorphism in CysLTR1 gene
We investigated a genetic variation in 5’ upstream region of CysLTR1 gene in 43 normal healthy controls (NCs) by direct sequencing and found 3 SNPs in CysLTR1 gene promoter. The transcription start site of the human CysLTR1 transcript was determined by 5’-RACE and three polymorphic sites: C-634T, A-475C, A-336G were identified from the transcriptional start site.
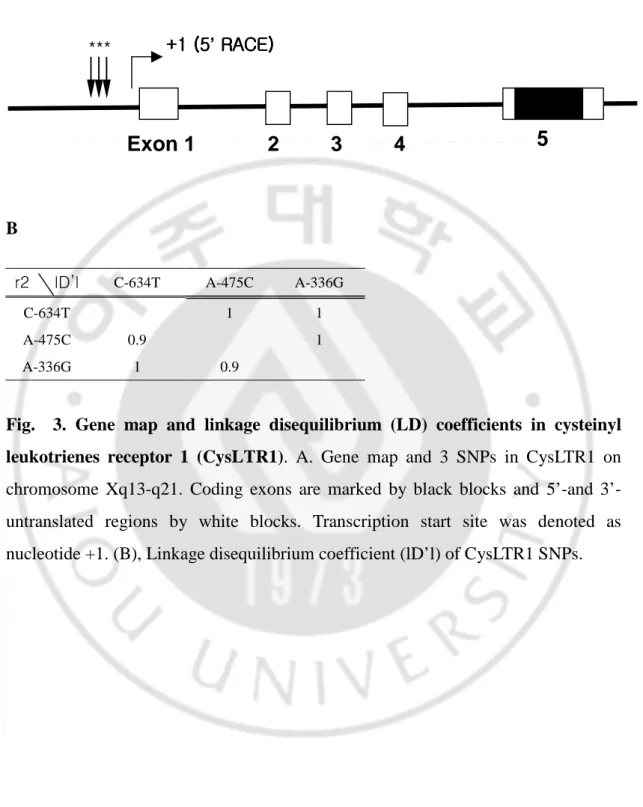
Next, we investigated genetic association study of three genetic polymorphisms of CysLTR1 in three groups of study subjects classified as AIA (n=105), ATA (n=111), and NCs (n=125) by direct sequencing, in which the 2466bp promoter region of CysLTR1 gene was included. Nucleotide position on (+1) is transcription start site determined by 5’-RACE and the sequence is shown in Fig. 2. A graphic overview of 3 SNPs identified in relation to the exon / intron structure of human CysLTR1 gene is shown in Fig. 3A. The 3 SNPs were in strong linkage disequilibrium (LD) with each other and the linkage disequilibrium
coefficients (1D’1) and r2 among the SNPs were calculatedfor each SNP (Fig. 3B).
B. Genetic association study of CysLTR1 genetic polymorphism
1. Clinical characteristics of the study subjects
One hundred and five ASA-intolerant asthmatics, 111 ASA-tolerant asthmatics and 125 normal healthy control subjects were genotyped at the three CysLTR1 SNPs,
1 CATTTGGGAA TGGGTGAATC ACTGCCTTCC CCATTGCTCA AGAACAGAAA ATTAGCTAAT 61 GAAAGAAACA GGTAGGGGAG CACAAAGAAG AACCACAGGA AACACCCTCT CTGCCACAGC 121 TATTTCGGTG CCAACTTGAA GCTCAAGGAT TGAAGGGGGC CTGCCCCTCC ACGCCTGTGG 181 GTATTTCTCA TCAGGTGGTA CGAGAGACTG AGAAAAGAAA TGACACAGAG ACAAAGTATA 241 GAGAAAGAAC AGTGGGCCCA GGGGACCGGC AGCCTCAGCA TGTGAGGACC TGCACCAGCG 301 CTGGTCTCTG AGTTCCCTCA GTATTTATTG ATCATTATTT TTACTATCTT GGCAAGGGGA 361 GTGTAGCAGA GCAACAGGTG GGGAGAAGGT CAGCAGGAAA ACGTGAGCAA AGGATCTGTA 421 TCATGCATAA ATTCAAGGAA AGGTACTGTG CCTGGATGTG CATGTAGGCC AGATTTATGT 481 TTCACTTTAT ACAAACACCT CAGTGTAGCA AAGAGTAACA GAGCAGTATT TCTGCCAGCA 541 TATCTCGCCT CTAGCCACAG GGCGGTTTTA TCCTATCTCA GAATAGAAAG AATGGGAATG 601 GTTGGCTTTA CAGGAGACAT TCCATTCCCA GGGAGGAGCA GGAGACAGAA GCCTTCCTCT 661 TATCTCAACT GCAAAGAGAC CTCCCTCTTT CACTACTCCT CCTCAGCAGA GACCCTTTAC 721 GGGTGTTGTG CTGGGGTATG GTATCAAAGC AGAAACAATT TTTCCTGGTA CAGATCAAAA 781 TGGAATTTCT TGTGTCTTCC TTTTCTACAT AGACACAGTA ACAATCTGAT CTCTCTTATC 841 CCCACAAAGG ATCTAGCTCC TTTCTCTTTT CTTCATTTCC TGAGTATGAA TTGCCCTTCC 901 ACTGTAGCTC AAAATGGCTG TAACATTTCA GACTGGAGGA AAAGTGATAA TTCACTCTGT 961 GTCTCACTTA TAAATTTTCC TGTAGATATT TGGGGATCCA AACTCAGTTG AAGAAATACT 1021 AGTTTGAATG TCAGTTTAGT TACTAAAACT CCCTAGGAGA GGATTAGAAA GTCTAGTCTT 1081 GAATCCAAAT GTTAACATCC CTTCAAATGT GGTTGGATAG CCTTTTCCTC CAACTATCAC 1141 CATCACCACC ACAATCCAGA ACTCTGTGAA AAGAGATAAA GATTCAAGGG CCTTAAAGCG 1201 ATTGTCCCAG GTTGGTTGTG AGGTAATTCT TGTACCTAAA GATACAAAAT TTGAGTAAAT 1261 GACTTTGTTC CTACAGCAGG TAGCAAGCCA GAAGGCTACT GCTGGGCTCA TATTTTACAA 1321 GTACCTAAAA CTAGTTGTTA GTTATGGGGC ATGTTTACAA AGTGTTAACA TCAACATATT 1381 TATGCCCATC TCCATATAGA ATCTTCTTAA TATAACTCAA GATAATTGAT GTAACAAATT 1441 CACATCATGA TCTTACCAAT TTGGTTTTTA ACATCAAAGT GCTGCCCCAG GCTTCAATCA 1501 GCACATACCA GACTGCTATG TCCCTTTTCT CCAGAAGCCA TATTGAGCAC CCCACAGCTC (* C-634 T)
1561 TAACACCTCA GAGCCTAGCC AAAGGCCAAG AACACTTGCC TGATCCCCCA AATTTCCAAC 1621 ACAGGACCAT TGACAGCAAG CCCAGTCATT CCAGATCATC TTCAGTGGGA CAAAAAAAGA 1681 AAACAACAGT TTTAATCCAA TGGAGGCAAT TTATTGTTAT GAAGATTTCA GGAAACAAAG (* A-475C)
1741 CTCAAAAAGG AACCAGAAAG GAAAAGGCTT TTTAGAATGT GGATAGAGCC ACAAGTGTCA 1801 TTAAGGAGAG AGAGAGAGAA ACGGAGAGAT GAAATTTATG TTACTTAAGA TCAAGTTTCA (* A-336 G)
1861 CATAATGCCA GTTATATTAG CATATACTGG CAGGATTATC TTTCCCCACC TACAAATAGT 1921 CTAATGACCT CACAGTCACA GAAATCACAG AGAACTAAGC TGAAGAGAGA ACACTCGTCC 1981 CTGCTTCCCA TCTTAGAGCA GCTGAATAAT TTCCTGAGAA TTCTATTCCT GAAGCTAGGA 2041 AGAAAAGTTT ATTTATACAT ACACGCAACC TGCAAGTCTC CAGTTTCTAT TCTTCCTTCC 2101 TCTTTGACCC TTCCCCTCCC CCACTTTGCA CCAGAGAAGT CAGACTCCGG GAGTGCTTTA (5’ RACE/*+1) 2161 ACAGTTTGAA GGCTAATCTG AAAGAGGAAG AAGAATCTGT ATATCTGTAT ATATTGGCTA ( EXON I)
2221 GCAAATGTGC CCTGCTCTCT CCCCTCTTAA AAATAGCAGC AACCCATCTT TGCAAAGAAG 2281 CTTGCCTATA GAGCAGGCAC TCTGTGAATG GACTGTGCTT TTACGACCCT ACAGGGTATC 2341 AAGATACTGT GCAGCTCGCC AACAAGGATT AATTGCAAGG ACTGGTAGAT CGAATTTACT 2401 GAAGACTTGG AGCTTGCTTC TGAGAACAAA CGCAAAAGGA CAGTAAACTG TGGACCTTGA 2461 AGTTAGCAGCG
Fig. 2. 5’-RACE sequence and 3 SNPs location on the 2466bp fragment of CysLTR1gene. Transcription start site as denoted as +1 found from the 5’-RACE and presented in bold letter and under line (_). Sequences of three genetic variants (C-634T, A-475C, and A-336G) in promoter region were represented as aterisk (*).
A B r2 lD’l C-634T A-475C A-336G C-634T 1 1 A-475C 0.9 1 A-336G 1 0.9
Fig. 3. Gene map and linkage disequilibrium (LD) coefficients in cysteinyl leukotrienes receptor 1 (CysLTR1). A. Gene map and 3 SNPs in CysLTR1 on chromosome Xq13-q21. Coding exons are marked by black blocks and 5’-and 3’- untranslated regions by white blocks. Transcription start site was denoted as nucleotide +1. (B), Linkage disequilibrium coefficient (lD’l) of CysLTR1 SNPs.
5
Exon 1 2 3 4
+1 +1 +1
+1 (5(5(5(5’ RACE) RACE) RACE) RACE) ***
C-634T, A-475C, and A-336G. The clinical characteristics of the study subjects are summarized in Table 1. There were no significant differences in the FEV1 % predicted, total IgE level, asthma duration and airway hyperresponsiveness to PC20 methacholine between the AIA and ATA groups (p=0.37, 0.20, 0.09, 0.22, respectively). There were no significant difference in mean age, prevalence of sex, atopy status among three groups (p>0.05). Between the AIA and ATA groups, there
were significant differences in percent in fall of FEV1 during lysine-aspirin
bronchoprovocation test (p<0.001). There were significant difference in the prevalence of nasal polyps between AIA and ATA, this results suggest that AIA patient (48%) were found to have significantly higher frequencies of nasal polyps than ATA patients (6.7%, p=0.003).
As the CysLTR1 gene is on the X chromosome, the distribution of the CysLTR1 promoter genotypes within male subjects deviated from Hardy-Weinberg equilibrium (p<0.01). All analyses were stratified by sex.
2. Genotype distribution of CysLTR1 3 SNPs
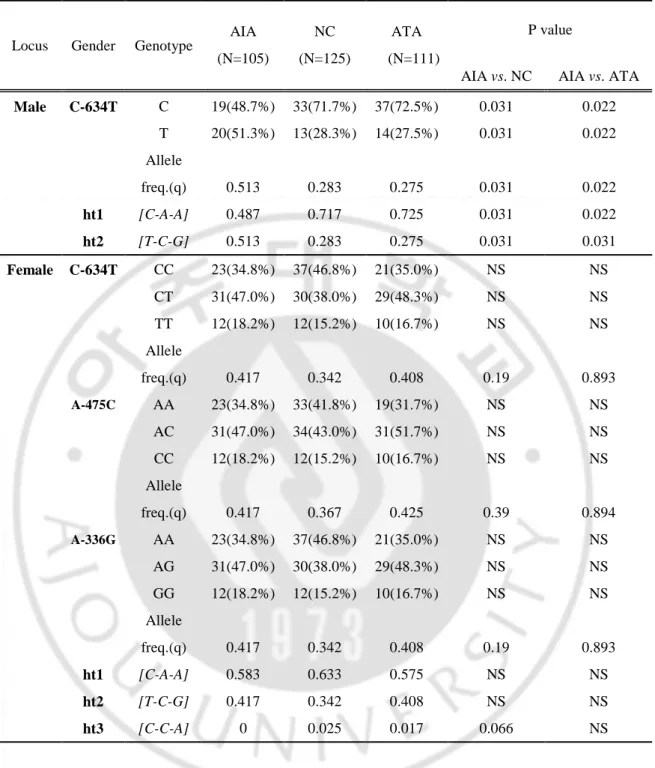
Next, we investigated a genetic association study of three genetic polymorphisms CysLTR1 in three groups of study subjects classified as AIA (n=105), ATA (n=111), and NCs (n=125). Allele and genotype frequencies of the 3 SNPs within the phenotypic groups are described in Table 2. Allele frequencies (q) were determined for each polymorphism. Male AIA patients were found to
have significantly higher frequencies of the minor alleles (T, C,G) of the CysLTR1 Table 1. Clinical characteristics of the study subjects
P value Characteristics AIA (n=105) NC (n=125) ATA
(n=111) AIA vs. NC AIA vs. ATA Age (year) 44.1 ± 13.4 45.5 ± 17.6 44.3 ± 14.9 NS NS
Sex (M) 39 (37.1%) 46 (36.8%) 51 (45.9%) NS NS
Atopy 53 (50.5%) 18(18.6%) 58 (55.2%) NS NS
Asthma duration (year) 6.7 ± 5.7 NA 5.3 ± 6.2 NS NS
FEV1 (%) 83.4 ± 22.9 NA 86.3 ± 23.1 NS NS
Fall of FEV1 (%) 30.0 ± 11.3 NA 4.5 ± 1.8 NS <0.001
PC20,methacholine
(mg/ml)
5.0 ± 8.6 NA 6.1 ± 9.9 NS NS
Log total IgE (IU/ml) 2.2 ± 0.54 1.9 ± 0.6 2.3 ± 0.6 0.001 NS
AR 82 (78.1%) NA 24 (85.7%) NS NS
PNS 56 (58.3%) NA 3 (50%) NS NS
Nasal polyps 48 (48.0%) NA 1 (6.7%) NS 0.003
AIA: ASA-intolerant asthma; NC: normal controls; ATA: ASA-tolerant asthma. N: number of patients; NA: not applicable; NS: not significant; M: male. Data are expressed as the mean ±SD
The methacholine provocative concentration producing a 20% fall in forced expiratory volume in 1 second (PC20-FEV1) was calculated as an index of bronchial hyperresponsiveness (BHR).
promoter SNPs than male control subjects (p=0.031 for AIA vs. NC; p=0.022 for AIA vs. ATA). In contrast, there were no significant differences in allele and genotype frequencies among the three groups within female subjects (p>0.05). Haplotypes of CysLTR1 gene were constructed by Bayesian algorithm with genotyped SNPs (Table 2) and 2 common haplotypes (frequency > 5%) were analyzed. Among males, AIA patients had a lower frequency of major haplotype, ht1 [C-A-A] (p=0.031, OR=0.36, 95%CI: 0.14~0.90 for AIA vs. NC; p=0.022, OR=0.34, 95%CI: 0.14~0.85 for AIA vs. ATA), and higher frequency of minor haplotype, ht2 [T-C-G] (p=0.031, OR=2.42, 95%CI: 0.98~5.98 in AIA vs. NC; p=0.031, (OR=2.57, 95%CI: 1.04~6.31 in AIA vs. ATA), than those in male control groups. Significant differences in the frequency of ht1 and ht2 were not found in female subjects. In addition, there were no significant differences in the haplotype distributions in female subjects by three alternative analysis models (co-dominant, dominant and recessive models).
3. Association analysis of AIA-associated quantitative phenotypes
Initially, AIA related phenotypes such as atopy, serum total IgE level, initial baseline FEV1 % predicted value and PC20 methacholine were evaluated for any association with the CysLTR1 C-634T SNP which is a tagging SNP of the 3 SNPs (Table 3). In female AIA patients, with mutant genotypes of CysLTR1 promoter polymorphisms showed higher total IgE levels (p=0.003), while no significant associations were found in male subjects. No significant associations were found between CysLTR1 polymorphisms and other phenotypes such as airway
Table 2. Genotype frequencies of the three SNPs in CysLTR1 promoter
Locus Gender Genotype
P value AIA (N=105) NC (N=125) ATA (N=111)
AIA vs. NC AIA vs. ATA
Male C-634T C 19(48.7%) 33(71.7%) 37(72.5%) 0.031 0.022 T 20(51.3%) 13(28.3%) 14(27.5%) 0.031 0.022 Allele freq.(q) 0.513 0.283 0.275 0.031 0.022 ht1 [C-A-A] 0.487 0.717 0.725 0.031 0.022 ht2 [T-C-G] 0.513 0.283 0.275 0.031 0.031 Female C-634T CC 23(34.8%) 37(46.8%) 21(35.0%) NS NS CT 31(47.0%) 30(38.0%) 29(48.3%) NS NS TT 12(18.2%) 12(15.2%) 10(16.7%) NS NS Allele freq.(q) 0.417 0.342 0.408 0.19 0.893 A-475C AA 23(34.8%) 33(41.8%) 19(31.7%) NS NS AC 31(47.0%) 34(43.0%) 31(51.7%) NS NS CC 12(18.2%) 12(15.2%) 10(16.7%) NS NS Allele freq.(q) 0.417 0.367 0.425 0.39 0.894 A-336G AA 23(34.8%) 37(46.8%) 21(35.0%) NS NS AG 31(47.0%) 30(38.0%) 29(48.3%) NS NS GG 12(18.2%) 12(15.2%) 10(16.7%) NS NS Allele freq.(q) 0.417 0.342 0.408 0.19 0.893 ht1 [C-A-A] 0.583 0.633 0.575 NS NS ht2 [T-C-G] 0.417 0.342 0.408 NS NS ht3 [C-C-A] 0 0.025 0.017 0.066 NS
*Each P value was calculated with co-dominant, dominant and recessive models. q: minor allele frequency; AIA: ASA-intolerant asthma; NC: normal controls; ATA: ASA-tolerant asthma; N: number of patients; NS: not significant. Values in bold indicate significant p value.
* C-634T is a tagging SNP of the three SNPs in CysLTR1 promoter. Significant associations (P<0.01) are printed in bold. P values are genotype-specific means ± SD.
Table 3. Association of CysLTR1 C-634T with atopy, total IgE, asthma duration, PC20
methacholine, FEV1 and fall of FEV1.
Gender Genotype Age Atopy
Log IgE (IU/ml) Asthma duration (year) PC20 methacholine (mg/ml) FEV1 (%) Fall of FEV1 (%) Male C 43.16 ± 12.9 11(57.9%) 2.26 ± 0.63 8.4 ± 6.1 4.56 ± 8.78 84. 85 ± 25.1 20.3 ± 10.4 T 36.1 ± 12.2 11(55.0%) 2.34 ± 0.57 5.5 ± 4.2 8.81 ± 15.3 76.79 ± 23.8 21.5 ± 13.0 p 0.087 0.86 0.670 0.095 0.268 0.309 0.769 Female CC + CT 47.7± 12.9 22(40.7%) 2.08 ± 0.47 6.5 ± 6.1 4.01 ± 7.05 86.7 ± 20.2 23.2 ± 11.7 TT 42.6 ± 13.7 9(75%) 2.56 ± 0.55 7.2 ± 5.6 3.76 ± 4.83 77.3 ± 28.5 19.6 ± 7.1 p 0.226 0.031 0.003 0.716 0.929 0.182 0.438
hyperresponsiveness to metahcholine and pulmonary function in both sexes.
4. Effect of 3 SNPs on transcriptional activity
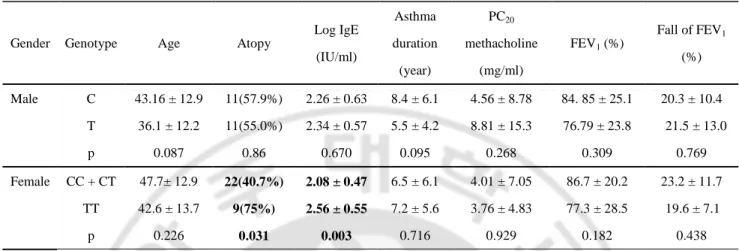
To explore the functional significance of the CysLTR1 promoter polymorphisms at the molecular level, promoter fragments containing all 2466bp CysLTR1 promoter haplotypes ht1 [C-A-A] and ht2 [T-C-G] were cloned into 5’ of the luciferase reporter gene within pGL3 basic vector. To perform this experiment, we screened several cell lines such as A549 cells, Hela cells, HL-60 cells, HUVEC, THP-1 cells, HEK-293T cells, U937 cells, Jurkat cells, and COS-7 cells by RT-PCR. Among them, endogenous expression of CysLTR1 was detected in HL-60 cells, Jurkat cells, U937cells, and A549 cells (Fig. 4). We performed transfection using lipofectaime into HL-60 and THP-1 cells and transfection efficiency of both cells was too low and therefore, we conducted transfection experiments in Jurkat cells, U937 cells, and A549 cells. Fig. 5A, 5B, and 5C summarized the data for the transient transfection and luciferase analysis in Jurkat cells, U939 cells, and A549 cells. The reporter activities were compared between two constructs containing either ht1 [C-A-A] or ht2 [T-C-G] at -634 bp, -475 bp, and -336 bp in the CysLTR1 promoter region (Fig. 5A, B and C). Luciferase activity was enhanced with the construct with minor haplotype, ht2 [T-C-G] (150% increases) compared to the construct with major haplotype ht1 [C-A-A].
5. Identification of unknown nuclear protein binding by EMSA
Fig. 4. Expression of CysLTR1 mRNA in various cell lines. Four micrograms
of total RNA was subjected to RT-PCR with the primers for CysLTR1. PCR products were separated on 1% agarose gel and stained with ethidium bromide. A: A549 cells; B: Hela cells; C: HL-60; D: HUVEC cells; E: THP-1 cells; F:
Fig. 5. Promoter activity assay of human CysLTR1 promoter constructs. Data
are mean values of independent mean ± SEM. (A) Transfection into Jurkat cell. Luciferase activity assay was performed nine times (in total, n=27). (B) Transfection into U937 cell (n=9) and (C) transfection into A549 cells (Human lung carcinoma, n=12). *P<0.05, **P<0.01, ***P<0.001 R L U 0 1 2 3 4 5 6 Basic ht1 [C-A-A] Ht2 [T-C-G] *** ** * A A A A 0 1 2 3 4 5 6 Basic ht1 [C-A-A] ht2 [T-C-G] * * B B B B R L U 0 1 2 3 4 5 6 7 ** ** *** Basi ht1[C-A-A] ht2 [T-C-G] C C C C R L U
SNPs regions, we searched transcription factor binding site (TFSEARCH, Searching Transcription Factor Binding Sites ver. 1.3.) and signal scan database (www. bimas.cit.nig.gov/molbio/signal), but this region has not putative transcription factor binding sites.
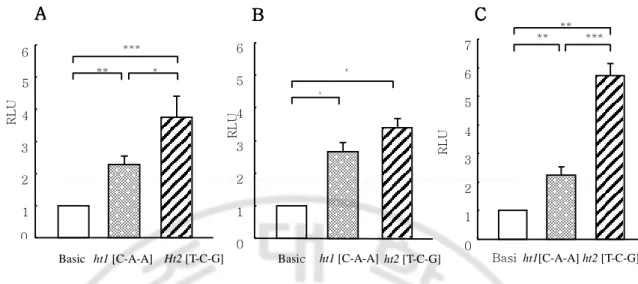
We found that a shifted band was noted on- 475A, -336A, and -336G of CysLTR1 promoter in nuclear extracts of IL-4 primed THP-1 cells (10ng/ml for 30min) and PMA (100ng/ml for 4h) stimulated U937 cells (Fig. 6A. and 6B.). To investigate whether the genetic variants create a transcription factor binding site, each SNPs containing double strands oligonucleotide probe( C-634T, 475C, A-336G) was synthesized and labeled with [α-32P]CTP as probe and incubated with each nuclear extracts of IL-4 primed THP-1 cells (10ng/ml for 30min) and PMA (100ng/ml for 4h) stimulated U937 cells. PMA stimulated U937 nuclear extract were represented in the formation of three specific protein-DNA complexes and contained nuclear proteins binding specifically to -475A and -336A and -336G region of CysLTR1 promoter. IL-4 stimulated THP-1 nuclear extracts revealed at least two major complexes. The formation of the band was inhibited completely by adding 50X excess of unlabeled - 475A oligonucleotide.
C. Characterization of CysLTR1 promoter in A549 cells
The finding of CysLTR1 expression in the A549 cells suggests that this cell line is a useful model system for studying the transcriptional regulation of human CysLTR1 gene. Various human CysLTR1 promoter-luciferase constructs were
1 2 3 -4 7 5 A 5 0 X -4 7 5 C 5 0 X N F -k B 5 0 X 1 2 -6 3 4 C 5 0 X -6 3 4 T 5 0 X N F -k B 5 0 X -3 3 6 A 5 0 X -3 3 6 G 5 0 X N F -k B 5 0 X -4 7 5 A 5 0 X -4 7 5 C 5 0 X N F -k B 5 0 X A B
Fig. 6. Unknown factor bind to a -475A, -336A, and -336G region in the CysLTR1 promoter. (A), EMSA with each nuclear extracts prepared from unstimulated line (1, 6, 11) and PMA (100ng/ml for 4hr) stimulated line (2-5, 7-10, and 12-15) of U937 cell lines. EMSA revealed at least three specific protein-DNA complexes. An unlabeled oligo with -475A and -336A, and -336G competed for bands 2 but not band 1 and 3. (B), EMSA with nuclear extracts prepared from IL-4 stimulated (10ng/ml for 30min) THP-1 cell lines and the revealed two protein-DNA complexes. The formation of the bands 1 was specific inhibited completely by -475A unlabeled oligo. The Lane 1, 6, 11 were represented probe only.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15
probe -634C -475A -336A - 634C -475A -336A Competitor -4 7 5 A 5 0 X -4 7 5 C 5 0 X N F -k B 5 0 X -3 3 6 A 5 0 X -3 3 6 G 5 0 X N F -k B 5 0 X -6 3 4 C 5 0 X -6 3 4 T 5 0 X N F -k B 5 0 X
transfected into the A549 cells to determine their promoter activities. The construct B567 (–256/+313)-Luc was found to have highest promoter activity in the transfected cells. The construct B981 (–670/+313)-Luc was found to have the
second promoter activity and further 3′ deletion of this construct [to generate the
B808 (–670/ +215)-Luc] could significantly abolish the promoter activity (Fig. 7A.), suggesting that the region within +215/+313 contains important transcriptional elememt(s) for the promoter function. IL-4 stimulation significantly increased luciferase activity in all tested construct (Fig.7B).
D. Transcriptional regulation of CysLTR1 gene by IL-4
1. Expression of CysLTR1
To investigate whether IL-4-increases at mRNA and protein levels of
CysLTR1 gene, analysis of CysLTR1 mRNA expression was measured by real-time RT-PCR, and CysLTR1 cell surface expression was determined by flow cytometric analysis. IL-4 stimulation increased CysLTR1 mRNA in a time-dependent fashion, with the highest increase was observed after 24 h of incubation (Fig. 8A). IL-4 stimulation caused a increased in surface receptor expression, and maximum response was observed at 24 h of incubation (Fig. 8B).
CysLTs promoted generation of Th2 cytokines such as IL-4, IL-5, IL-13 and B 0.0 5.0 10.0 15.0 20.0 25.0 30.0 B2400 B981 B808 B567 fo ld i n d u c ti o n Control IL-4 -2158
A Human CysLTR1 5’-flanking region
0 2 4 6 8 1 1
Relative promoter activity (fold of increase)
-1268 -670 -256 +1 +205 +313 pGL3 basic vector B808 (-670/+205) B567 (-256/+313) B981 (-670/+313) B1570 (-1268/+313) B2400 (-2158/+313) Luc Luc Luc Luc
**
*
*
*
*
*
Luc Luc*
*
Fig. 7. Deletion analysis of the human CysLTR1 promoter. (A), A549 cells were
transfected by lipofectamin with five CysLTR1 promoter luciferase based reporter constructs as well as promoter- less vector (pGL3 basic), and allowed to recovery for 48hr. (B), a exposed to IL-4 or vehicle for 6h. Analysis of reporter gene expression using luminometry. Data are the mean ± SEM of six experiments; asterisk indicates (**) p<0.001and (*) p<0.01 versus promoterless vector (pGL3 basic vector). Numbers underneath refer to nucleotide up stream of the transcription start site.
0 0.5 1 1.5 2 2.5 3 Cellonly 2hr 4hr 6hr 8hr 24hr m R N A ( fo ld ) Hours A A A A BBBB
Fig. 8. CysLT1R mRNA and protein expression. (A), IL-4 increased CysLTR1 mRNA, A549 cells were stimulated with IL-4 (10 ng/ml) for up to 24 h and CysLTR1 mRNA expression was measured by realtime RT-PCR analysis. Results are normalized to internal control (β-actin expression) and presented as fold increase from control values (treated cells vs. vehicle). The means SD of three different samples are shown. (B), Flow cytometric analysis of CysLTR1 surface expression. A549 cells were stimulated with IL-4 (10 ng/ml) for 18 h and labeled with anti-CysLTR1 antibody or with isotype-matched control. Results of a single experiment, representative of at least three are presented. Dotted line represents isotype control antibody. Solid thick and thin lines represent labeling of medium- and IL-4-treated cells, respectively.
those cytokines also enhanced generation of CysLTs in macrophage, eosinophils, and monocytes by inducing the expression of CysLTR1 (Peters-Golden and Sampson, 2003; Thivierge and Pleszczynski, 2001; Thivierge and Rola-Pleszczynski, 2000; Chibana and Fukuda, 2003). We investigated whether IL-4 possess effect on transcriptional induction of CysLTR1 promoter, classified according to major haplotype, ht1 [C-A-A], and minor haplotype, ht2 [T-C-G]. There were no significant changes observed in the promoter activities in major haplotype, ht1 [C-A-A] by IL-4. In contrast, minor allele ht2 [T-C-G] had significantly higher promoter activity than untreated cells (p<0.001, Fig. 9.). We next determined to the effects of Th2 cytokine (IL-4), dexamethasone, and CysLTR1 specific antagonist (MK-571) on the CysLTR1 promoter activity in A549 cells.
E. Effect of leukotriene modifier on the transcription regulation by IL-4
Corticosteroids represent one of the most widely used and the most effective anti-inflammatory treatment of immune and anti-inflammatory diseases, including persistent asthma of all degree severity (Boumpas, 1993). Their mechanism of action involves regulation of gene transcription and RNA stability. CysLTs have been identified as an important brochoconstrictor in asthma, particularly in AIA and CysLTR1 selective antagonists could block bronchoconstriction induced by lysine aspirin in some patient with AIA (Dahlen B et al, 1993). To investigate whether MK-571 and dexamethasone possess an inhibitory effect on transcriptional induction of CysLTR1 induced by IL-4, we compared transcription activities of
Fig. 9. Effect of dexamethasone and MK-571 on CysLTR1 promoter
activity. Luciferase reporter activities were in A549 cells transfected with
CysLTR1 promoter construct (B2400). After total transfection time of 48h, the cells were incubated with various concentrations of dexamethasone and MK-571 for 24hr. The results are presented as the mean ± SD; n=9.
Drug dose 0.0 5.0 10.0 15.0 20.0 25.0 30.0 RL U Dex (M): - 10-9 10-8 10-7 10-6 - - - - MK-571 (M): - - - - - 10-9 10-8 10-7 10-6 WT [C-A-A] MT [T-C-G]
CysLTR1 promoter gene between major allele ht1 [C-A-A] and minor allele ht2
[T-C-G]. Compare to untreated cells, maximal inhibitions was observed at 10-8M of
dexamethasone and 10-7M of MK-571 (Fig. 9). We next determined the effects of
Th2 cytokine (IL-4), dexamethasone, and CysLTR1 specific antagonist (MK-571) on the CysLTR1 promoter activity in A549 cells. Significant increase of promoter activity was noted in mutant allele ht2 [T-C-G] (p=0.008), not in major allele ht1
[C-A-A], which was inhibited by 10-7M of MK-571 pretreatments (p=0.004, Fig. 10).
However, dexamethasone pretreatments increased promoter activity in both mutant allele ht2 [T-C-G] and major allele ht1 [C-A-A] (p<0.001).
F. Effect of dexamethasone on CysLTR1 mRNA expression in A549 cells
To investigate whether to assess the effect of dexamethasone on CysLTR1 expression in A549 cells, cells were incubated with fresh medium in presence or
absence of various dexamathasone from 10-9M to10-6M (Fig. 11). Treatment of A549
cells with various concentration dexamethasone increased CysLTR1 mRNA expression compared to untreated cells. Maximum enhancement was observed