Prevalence of tumor BRCA1 and BRCA2 dysfunction in unselected patients with ovarian cancer
Roshni D. Kalachand, MBBCh
1, Ciaran O’Riain, MBBCh
2, Sinead Toomey, PhD
1, Aoife Carr, BSc
1, Kirsten M. Timms, PhD
3, Sharon O’Toole, PhD
4, Stephen Madden, PhD
5, Mark Bates, PhD
4, John J. O’Leary, MBBCh
2, Noreen Gleeson, MD, MRCOG
4, Dearbhaile O’Donnell, MBBCh
6, Liam Grogan, MBBCh
7, Oscar Breathnach, MBBCh
7, Angela Farrelly, PhD
1, Britta Stordal, PhD
8, Bryan T. Hennessy, MD
1,7,91
Medical Oncology Group, Department of Molecular Medicine, Royal College of Surgeons in Ireland, Beaumont Hospital,
2Department of Histopathology, Trinity College Dublin, Central Pathology Laboratory, St. James’s Hospital, Dublin, Ireland;
3Myriad Genetics, Inc., Salt Lake City, UT, USA;
4Department of Obstetrics and Gynaecology, Trinity College Dublin, Trinity Centre for Health Sciences, St. James’s Hospital,
5Data Science Centre, Royal College of Surgeons in Ireland, Beaux Lane House,
6Department of Clinical Medicine, Trinity College Dublin, St. James’s Hospital,
7Department of Medical Oncology, Beaumont Hospital, Dublin, Ireland;
8Department of Natural Sciences, Middlesex University, Hendon, London, United Kingdom;
9
Our Lady of Lourdes Hospital, Drogheda, Ireland
Objective
The therapeutic benefits of poly(ADP-ribose) polymerase inhibitors highlight the need to evaluate BRCA1/2 defects in tubal/ovarian cancer (OC). We sought to determine the pattern and disease characteristics associated with tumor BRCA1/2 mutations and BRCA1 methylation in women with OC.
Methods
We obtained 111 OC specimens from 2 university hospitals and assessed BRCA1/2 mutations and BRCA1 methylation in tumor DNA. The frequency and pattern of BRCA1/2 defects were examined. Associations between patient/disease characteristics and BRCA1/2 defects were ascertained (Fisher’s exact test). Platinum-free interval (PFI), progression- free survival (PFS), and overall survival (OS) based on the underlying BRCA1/2 defect were determined (Kaplan-Meier analysis [log-rank test]).
Results
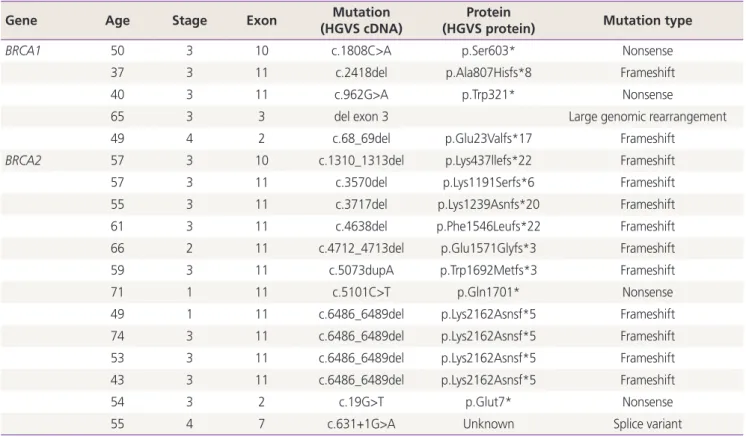
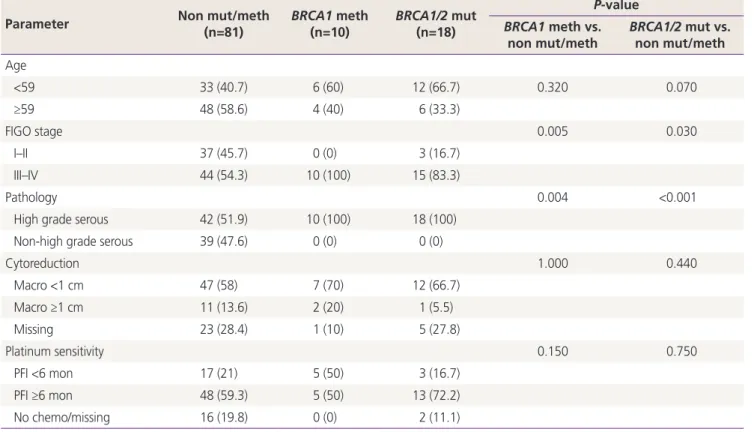
We observed a BRCA1/2 dysfunction rate of 40% (28/70) in high-grade serous tubal/ovarian cancer (HGSC), including 14.3% BRCA1 methylation (n=10), 7.1% BRCA1 mutation (n=5), and 18.6% BRCA2 mutation (n=13). Defects in BRCA1/2 genes were associated with stage III/IV HGSC (BRCA1 methylation: P=0.005 [stage III/IV] and P=0.004 [HGSC];
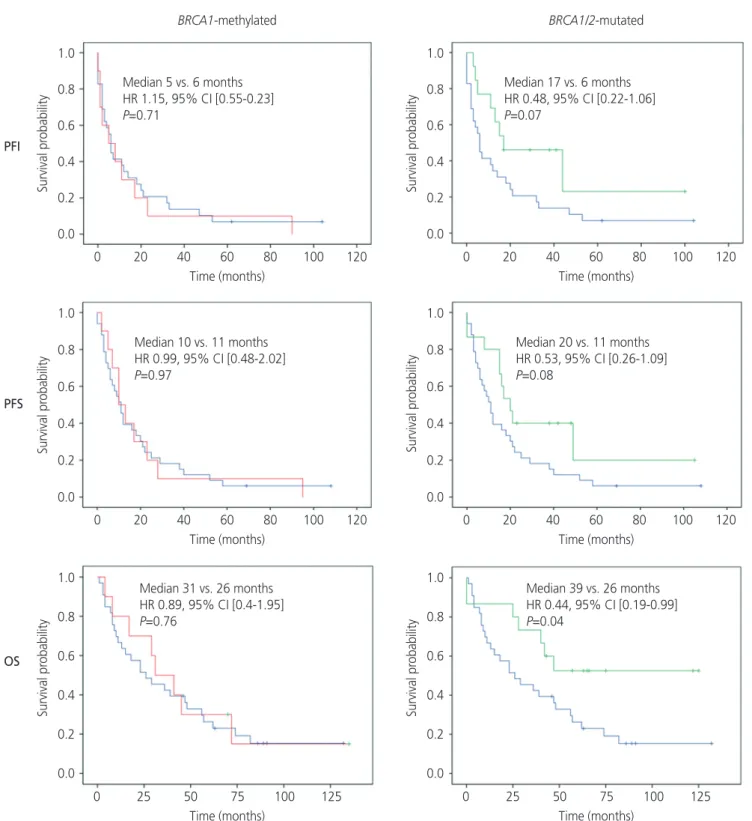
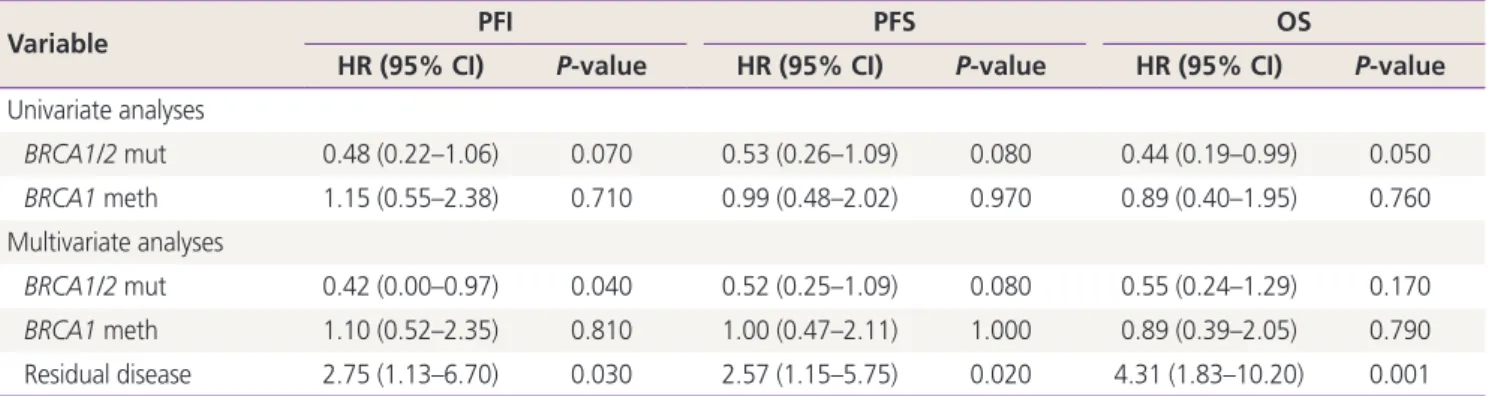
BRCA1/2 mutation: P=0.03 [stage III/IV] and P<0.001 [HGSC]). Patients with BRCA1/2-mutated cancers showed improved OS (hazard ratio [HR], 0.65; 95% confidence interval [CI], 0.43–0.99; P=0.045) and a trend toward improved PFI (HR, 0.48; 95% CI, 0.22–1.06; P=0.07) and PFS (HR, 0.72; 95% CI, 0.51–1.03; P=0.07). No survival differences were observed between BRCA1-methylated and BRCA1/2 wild-type non-BRCA1-methylated cancers.
Conclusion
We observed a high tumor BRCA1/2 dysfunction rate in HGSC with a unique predominance of BRCA2 over BRCA1 mutations. While BRCA1/2 mutations conferred survival benefits in OC, no such association was observed with BRCA1 methylation.
Keywords: Ovarian cancer; BRCA1 methylation; BRCA1 mutation; BRCA2 mutation
Received: 2020.02.04. Revised: 2020.05.03. Accepted: 2020.05.06.
Corresponding author: Roshni Kalachand, MBBCh
Medical Oncology Group, Department of Molecular Medicine, Royal College of Surgeons in Ireland, Smurfit Building, Beaumont Hospital, Dublin 9, Ireland
E-mail: rkalachand@rcsi.ie
https://orcid.org/0000-0003-3060-7877
Articles published in Obstet Gynecol Sci are open-access, distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.
org/licenses/by-nc/3.0/) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Copyright © 2020 Korean Society of Obstetrics and Gynecology https://doi.org/10.5468/ogs.20033
pISSN 2287-8572 · eISSN 2287-8580
Introduction
Poly (ADP-ribose) polymerase inhibitors (PARPi) exhibit potent activity in germline BRCA1/2-mutated platinum-sensitive relapsed high-grade serous tubal/ovarian cancer (HGSC). In phase III clinical trials, maintenance therapy with PARPi was associated with a 73% reduction in the risk for disease pro- gression or death as compared to placebo [1]. PARPi target the homologous recombination DNA repair defect (HRD) conferred by BRCA1/2 mutations, leading to tumor genomic instability and cell death. While germline BRCA1/2 mutations are detected in 15% HGSCs, genomic and functional data suggest the presence of HRD in approximately 50% HGSC [2]. The identification and validation of other HRD-associated biomarkers in sporadic tubal/ovarian cancer (OC) (hereafter referred to as OC) are crucial to potentially expand the num- ber of women with OC who could benefit from DNA repair- targeting agents such as PARPi.
Somatic BRCA1/2 mutations have been identified in 4–6.4% of HGSCs, wherein they account for 14.2% of HRD cases [2,3]. Evidence suggests that the clinical benefit from PARPi in patients with somatic BRCA1/2-mutated HGSC is similar to that observed in those with germline BRCA1/2- mutated disease. In the phase III NOVA clinical trial, 19.7%
(n=40) of BRCA1/2 mutations were classified as somatic [1]. The median progression-free survival (PFS) associated with niraparib as compared to that with placebo (20.9 vs.
11 months, hazard ratio [HR], 0.27; 95% confidence inter- val [CI], 0.08–0.9; P=0.02) in this subgroup was consistent with the value reported for patients with germline BRCA1/2- mutated disease (21 vs. 5.5 months, HR, 0.27; 95% CI, 0.17–0.41; P<0.001) [1].
BRCA1 promoter methylation has been identified as a potential biomarker of response to the PARPi rucaparib [4].
BRCA1-methylated tumors are negative for BRCA1 gene and protein expression, suggestive of a resultant HRD phenotype [5,6]. In addition, BRCA1-mutated and BRCA1-methylated OCs display similar gene signatures, as detected using gene expression and copy number analyses [7]. In the phase II open label ARIEL-2 study, 12/19 (63%) relapsed platinum- sensitive BRCA1-methylated OC patients responded to ru- caparib as compared to an 80% response rate reported in BRCA1/2-mutated OC patients [4]. This early data suggest the potential role of BRCA1 methylation as a biomarker of response to PARPi.
Considering the benefit of PARPi in BRCA1/2 dysfunctional OC and the ongoing development of other agents targeting DNA repair, the knowledge of the prevalence and pattern of BRCA1/2 gene aberrations within an OC population is imper- ative. The use of tumor tissues offers the advantage of iden- tifying additional potential somatic biomarkers of response to PARPi as compared to germline mutation testing alone.
This information may serve as a guide to drug approval strat- egies for novel DNA repair targeting drugs at a national level, as the distribution of BRCA1/2 mutations varies between populations [8]. In Ireland, the frequency of BRCA1/2 gene aberrations in OC is yet to be examined. At the time of this study, genetic testing for BRCA1/2 mutations in OC in Ireland was carried out on the basis of clinical risk algorithms in a clinician-dependent manner.
Here, we sought to assess the BRCA1/2 gene profile in a cohort of Irish women with OC by determining the frequen- cies of BRCA1/2 mutations and BRCA1 methylation in tumors and their association with clinical characteristics and survival.
Materials and methods
1. Sample and data collection
We selected 111 patients with OC treated at 2 university teaching hospitals (including a national tertiary referral gy- necologic oncology unit) between 2005 and 2013. All histo- logical subtypes, stages, and grades were included to allow accurate assessment of BRCA1/2-mutated and BRCA1-meth- ylated profiles. Borderline tumors were excluded. In total, 100 patients were retrospectively included from a prospective clinically annotated Discovary bioresource (St. James’s Hospi- tal) after receiving ethical approval for this study (reference 2009/29/01). Patients provided written informed consents prior to specimen collection. Within this bioresource, all pa- tients with epithelial OC with available and adequate tumor tissues (>30% neoplastic cell content [NCC]) were included.
Eleven samples were obtained from the Beaumont Hospital
Pathology Department after receiving approval from the
hospital’s ethics committee (REC reference 12/02). Clinical
data for these patients were retrospectively obtained through
medical records. Patients recruited through both bioresources
presented either to the outpatient department or as direct
inpatient referrals. All survival data were updated to February
15, 2017. A pathologist specializing in gynecological cancers
reviewed fresh-frozen paraffin-embedded (FFPE) tumor speci- mens for histology and NCC (as per the 2014 World Health Organization Classification). All specimens were obtained prior to chemotherapy (either at primary debulking surgery or peritoneal biopsy). Specimens with less than 30% NCC (n=20) were macrodissected prior to DNA extraction. The majority of the specimens had over 60% NCC.
2. Assessment of tumor BRCA1/2 defects
The DNA was extracted from FFPE tumor samples using the QIAamp DNA FFPE Tissue kit (Qiagen, Venlo, The Nether- lands) according to the manufacturer’s protocol, and quanti- fied using the dsDNA BR assay kit (Qubit, London, UK) as per the manufacturer’s instructions.
BRCA1 methylation status was assessed using the Methyl- Profiler DNA Methylation polymerase chain reaction (PCR) Array System (SABiosciences, Valencia, CA, USA) following the manufacturer’s protocol. In brief, DNA methylation-sen- sitive and methylation-dependent restriction enzymes were used to selectively digest non-methylated or methylated ge- nomic DNA, respectively. After digestion, DNA samples were subjected to real-time PCR using primers flanking the regions of interest. The relative concentrations of differentially meth- ylated DNA were determined by comparing the amount of each digest with that of a mock digest. A cutoff value of 10% methylation was used to define the methylation status of samples.
BRCA1 and BRCA2 genes were sequenced using the Tumor BRACAnalysis CDx assay (Myriad Genetics, Munich, Germany and Salt Lake City, UT, USA), as previously described [9]. Only deleterious or suspected deleterious mutations were included in analyses (as per the previously defined criteria [10]). Germ- line or somatic mutation status was not assessed, owing to the restrictions imposed by patients’ informed consent.
3. Statistical analysis
Statistical analysis was performed using SPSS
®version 21.0 software. BRCA1/2 mutations and BRCA1 methylation were associated to the following variables: patient age, histol- ogy, International Federation of Gynecology and Obstetrics (FIGO) stage, degree of surgical cytoreduction, and platinum sensitivity using the Fisher’s exact test. Survival analyses were carried out for platinum-free interval (PFI), PFS, and overall survival (OS) to compare patients with BRCA1/2-mutated disease or BRCA1-methylated disease with patients carry-
ing BRCA1/2 wild-type non-BRCA1-methylated (hereafter referred to as BRCA1/2-intact) tumors. PFI was defined as the interval between completion of chemotherapy and disease recurrence (as defined by the CA125/RECIST criteria), death, or date of last follow-up, whichever occurred first. PFS was defined as the interval between first surgical debulking or diagnostic biopsy (for patients receiving adjuvant or neoad- juvant chemotherapy, respectively) and disease recurrence
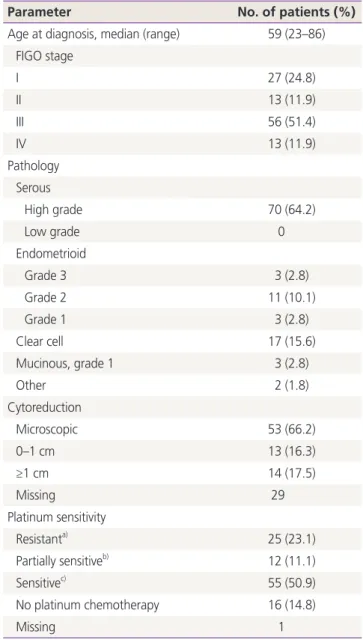
Table 1. Patient and disease characteristics
Parameter No. of patients (%)
Age at diagnosis, median (range) 59 (23–86) FIGO stage
I 27 (24.8)
II 13 (11.9)
III 56 (51.4)
IV 13 (11.9)
Pathology Serous
High grade 70 (64.2)
Low grade 0
Endometrioid
Grade 3 3 (2.8)
Grade 2 11 (10.1)
Grade 1 3 (2.8)
Clear cell 17 (15.6)
Mucinous, grade 1 3 (2.8)
Other 2 (1.8)
Cytoreduction
Microscopic 53 (66.2)
0–1 cm 13 (16.3)
≥1 cm 14 (17.5)
Missing 29
Platinum sensitivity
Resistant
a)25 (23.1)
Partially sensitive
b)12 (11.1)
Sensitive
c)55 (50.9)
No platinum chemotherapy 16 (14.8)
Missing 1
Percentages reflect percentage of total non-missing data.
FIGO, International Federation of Gynecology and Obstetrics; PFI, platinum-free interval.
a)