Expression of MicroRNA-221 in Korean Patients with Multiple Myeloma
전체 글
수치
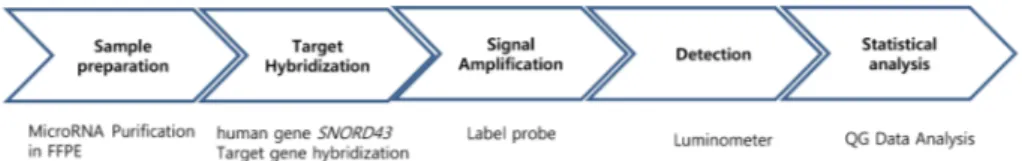

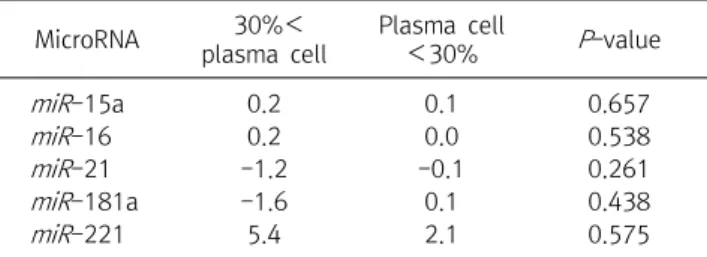
관련 문서
In addition, the expression of REDD1 was increased when Huh7 cells were treated with PA, and overexpression of REDD1 affected cell survival and lipid
A and E, In control group, a small amount of new bone was observed at the margin of bone defect (40×); B and F, In experimental group 1, a large amount of new bone was formed
mRNA expression of osteonectin, Runx2 and BSP(bone sialoprotein) in MC3T3-E1 cells cultured for 24 hours on 4 different titanium surfaces.. The mRNA were analyzed
This study was conducted under the hypothesis that a group art therapy program may increase emotional expression and decrease life event stress in patients with
Results : The expression of p21 was increased in boderline serous tumor and serous cystadenocarcinoma in contrast to benign serous tumors. The expression of
Results: In this research, in the group with fibromyalgia patients group, systemic lupus erythematosus patients group and without systemic autoimmune
▪ When we control the potential of the working electrode with respect to the reference, it is equivalent to controlling the energy of the electrons within the
4 > Effects of Taro on COX-2 expression and iNOS expression(hot water) in human thyroid cancer cells. The cells were pretreated for 48hours with either