Constitutive Expression of Defense Genes in Transgenic Arabidopsis Overproducing Methyl Jasmonate
Choonkyun Jung, Seoung Hyun Lyou, Yeon Jong Koo, Jong Tae Song, Yang Do Choi and Jong-Joo Cheong*
School of Agricultural Biotechnology, Seoul National University, Suwon 441-744, Korea Received March 21, 2003; Accepted May 12, 2003
Methyl jasmonate modulates diverse developmental processes and defense responses in plants, acting as an important cellular regulator. Formation of this volatile is catalyzed by a jasmonic acid carboxyl methyltransferase (JMT). Expression of various defense genes in transgenic Arabidopsis overexpressing the JMT gene, thus containing elevated level of endogenous methyl jasmonate, was examined.
Constitutive expression of various defense genes including those for defensin, pathogenesis-related proteins, and oxidative stress-resistant enzymes was observed through Northern and dot blot analyses.
The transgenic plants exhibited enhanced resistance to a non-host virulent bacterial pathogen, Pseudomonas syringae pv tomato DC3000. Taken together with previous reports, our data imply that genetic introduction of MeJA-producing gene could be used to achieve fortified resistance of plants against various pathogens.
Key words : Arabidopsis, defense genes, jasmonic acid carboxyl methyltransferase, methyl jasmonate, oxidative stress.
Plants exert a variety of defense mechanisms in response to pathogen attack as a consequence of transcriptional activation of defense-related genes. Accumulation of defense gene transcripts generally commences within minutes to hours around the infection sites, and several hours or days later at distant sites over the whole plant. In many local and systemic responses of plants, a large group of PR proteins are synthesized in high amounts to display a broad spectrum of antimicrobial activity.1) Through intensive studies, numerous genes involved in these processes have been identified.2)
Signal transducers such as jasmonates, salicylic acid, and hydrogen peroxide mediate the activation of plant defense genes.3,4) For instance, jasmonates trigger salicylic acid- independent defense mechanisms, activating the genes encoding toxic proteins such as defensin5) and thionin.6,7) Expression of the defensin gene PDF1.2 is responsible for the resistance against non-host fungal pathogens, Botrytis cinerea,5,8,9) Aternaria brassicicola,9) and Pythium mastophorum.10) In addition, the induced systemic resistance (ISR) response triggered by nonpathogenic rhizobacteria is also mediated by jasmonates.11)
Jasmonates, collectively referring to MeJA and its free acid JA, have been recognized as important cellular regulators. In
addition to the role(s) in defense responses against a group of pathogens12) and insect-driven wounding,13) jasmonates activate diverse developmental processes such as seed germination, flower and fruit development, leaf abscission, and senescence.14,15) Moreover, accumulating evidences suggest that jasmonates have a role in plant resistance against various environmental stresses such as drought, low temperature, osmosis, and high salt concentration.16)
Jasmonates are synthesized in plants via the octadecanoid pathway.14,17) At the end of this pathway, JA is catabolized further to form its volatile counterpart MeJA by a S-adenosyl- L-methionine: jasmonic acid carboxyl methyltransferase (JMT) activity.18) In the transgenic Arabidopsis overproducing the JMT gene and thus containing elevated level of endogenous MeJA concentration, various jasmonate- responsive genes were constitutively expressed in the absence of wounding or jasmonate treatment. This finding suggests that MeJA formation is a critical control point for jasmonate- regulated plant responses. Importance of JMT gene activation in the MeJA-induced defense response was demonstrated by the illustration that the transgenic Arabidopsis exhibited enhanced resistance to the virulent fungal pathogen Botrytis cinerea.18)
In this study, we observed that the transgenic Arabidopsis plants overexpressing the JMT gene constitutively express various defense genes. Furthermore, we found that the transgenic plants exhibited enhanced resistance to a bacterial pathogen, Pseudomonas syringae pv tomato DC3000. These results indicate that MeJA mediates plant defense responses against a broad spectrum of pathogens including fungi and bacteria. Thus, genetic introduction of the MeJA-producing
*Corresponding author
Tel: 82-31-290-2647, Fax: 82-31-293-8608 E-mail: cheongjj@snu.ac.kr
Abbreviations: AOS, allene oxide synthase; cfu, colony-forming unit;
EST, expressed sequence tag; JMT, S-adenosyl-L-methionine: jasmonic acid carboxyl methyltransferase; MeJA, methyl jasmonate; JA, jasmonic acid; PCR, polymerase chain reaction; PR proteins, pathogenesis-related proteins.
gene could be a way to achieve a fortified disease resistance of plants.
Materials and Methods
Plant materials. Arabidopsis thaliana (ecotype Columbia) plant was used as a wild-type control. For the generation of transgenic plants,18) a full-length JMT cDNA was inserted into the pBI121 (Clontech, USA) in sense orientation at downstream of the cauliflower mosaic virus 35S (CaMV 35S) promoter. The construct was transformed into Arabidopsis (ecotype Columbia) following the Agrobacterium-mediated floral dip transformation procedure. A T3 line of the transgenic Arabidopsis was prepared from the T2 plants previously described.18) All plants were sown in potting soil and grown at 21-23oC and 60% relative humidity under a cycle of white light at 500µmol · m−2· sec−1 (16 h) and darkness (8 h).
RNA isolation and Nothern blot analysis. Total RNAs were isolated from rosette leaves of individual plants using the ConcertTM Plant RNA Purification Reagent (Invitrogen, USA) and the RNeasy Mini Kit (Qiagen, USA). For Northern blot analysis, 10µg of total RNA was electrophoresed on 1.5%
formaldehyde agarose gel and blotted onto nylon membranes.
Loading of equal amount of RNA was confirmed by staining the gel with ethidium bromide. DNA probes for JMT and PDF1.2 transcripts were obtained as described.18)
Dot blot analysis. Expressed sequence tags (ESTs) of selected genes were obtained from The Arabidopsis Biological Resource Center (ABRC) of The Arabidopsis Information Resource (TAIR) and used as DNA probes (Table 1). Inserts in the ESTs were amplified by PCR and blotted on a Zeta-Probe membrane (Bio-Rad, USA). DNA probes for thaumatin-like protein mRNA (Genbank accession number M90510), chlorophyll a/b-binding protein mRNA (BT0000726), and 25S ribosomal RNA (X52320) were amplified from a home-made Arabidopsis cDNA library by PCR. A appropriate primers were designed based on the
sequences deposited on the Genbank database.
Four micrograms of total RNA was labeled with DIG-11- dUTP (Roche) by reverse transcription using Superscript II reverse transcriptase and oliogo(dT)15. The DIG-11-dUTP- labeled probe was purified using Sephadex G-50 column (Sigma, USA), and hybridized to a dot blot membrane at 42oC overnight. After hybridization, the membrane was washed twice for 10 min with pre-washing mixture of 2× SSC (sodium saline citrate) and 1% SDS (sodium dodecyl sulfate), then twice at 42oC for 10 min with washing solution containing 1× SSC and 0.1% SDS. After the post- hybridization washes, the membrane was washed again briefly with 1× TBST (Tris-buffered saline with 0.5% Tween 20), blocked with 5% skim milk at room temperature for 1 h, and treated with anti-DIG-POD-labeled antibody (1 : 5000) (Amersham Bioscience) for 1 h. After washing the membrane four times with 1× TBST for 10 min, hybridization signal on the membrane was detected by enhanced chemi-luminescence (Pierce).
Pathogen-resistance test. For the bacterial pathogen- resistance test, 7 week-old Arabidopsis plants were infected with the virulent pathogen Pseudomonas syringae pv tomato DC3000. Bacterial suspension (1× 108 cfu · ml−1) was sprayed onto the plants. The plants were incubated at 25oC and 100% relative humidity for 1 day, and transferred into a growth chamber maintained at 23oC and 50% relative humidity. The infected rosette leaves were harvested, weighed, rinsed thoroughly with sterile water, and homogenized in 10 mM MgSO4. Appropriate dilutions were plated onto King’s medium B agar. After incubation at 28oC for 1 or 2 days, the number of colonies on the medium was counted.
Results
Transgenic Arabidopsis overexpressing JMT. As described previously,18) the transgenic Arabidopsis plant used in this study integrates a full-length JMT cDNA (1.5 kb) fused Table 1. Expressed sequence tags (ESTs) used as DNA probes in this study.
ABRC stock numbera Associated locib Descriptionc
201H10 AT4G33720 PR-1
91B11 AT3G57260 β-1,3-glucanase (PR-2)
92G1 AT3G12500 Basic chitinase (PR-3)
245P5 AT3G04720 Hevein-like protein (PR-4)
209K10 AT2G43150 Extensin (Hrgp)
F10H11T7 AT2G37130 Peroxidase
178G17 AT2G28190 Co/Zn superoxide dismutase
179P3 AT1G45145 Thioredoxin
248H4 AT4G02520 Glutathione S-transferase
aArabidopsis ESTs were obtained from The Arabidopsis Biological Resource Center (ABRC) of The Arabidopsis Information Resource (TAIR).
bAssociated chromosomal loci of the corresponding genes are described with Arabidopsis Genome Initiative (AGI) numbers.
cDescription of the gene products is based on the functional definitions of The Institute for Genomic Research (TIGR).
with the CaMV35S promoter in the genome. We obtained T3 plants, and tested the expression of JMT gene in these plants by Nothern blot analysis. The transgenic plants contained high level of JMT transcripts (Fig. 1). In addition, expression of jasmonate-responsive genes such as PDF1.2 was elevated in the absence of wounding or external jasmonate treatment.
Expression levels of defense genes. Transcript levels of a set of defense genes in the transgenic Arabidopsis were determined by dot blot analysis and compared with those in the wild-type plants. Expression of various defense genes that encode β-1,3-glucanase (PR-2), basic chitinase (PR-3), hevein-like protein (PR-4), thaumatin-like protein (PR-5), and extensin (Hrgp) were significantly elevated in the transgenic plants (Fig 2). Oxidative stress-related genes coding for peroxidase, copper/zinc superoxide dismutase, and thioredoxin also showed elevated levels of transcripts in the transgenic Arabidopsis. Glutathione S-transferase gene was also constitutively expressed in the transgenic plants. By
contrast, chlorophyll a/b-binding protein gene showed significantly reduced level of transcription. Salicylic acid- responsive gene PR1 was not induced in these plants (data not shown). 25S ribosomal DNA used as a control showed a constant expression level in all plants tested.
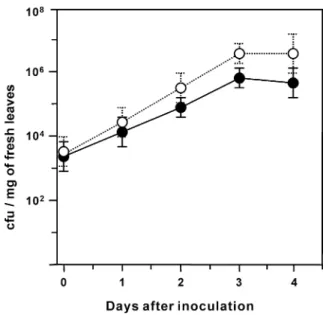
Resistance to bacterial pathogen. The transgenic Arabidopsis overexpressing the JMT gene exhibited significantly high degree of resistance against the infection with virulent pathogen Pseudomonas syringae pv tomato DC3000. On the transgenic plants inoculated with 1× 108 cfu
· ml−1 of the bacteria, growth rate of the bacteria was reduced significantly (Fig. 3). Three or four days after the infection, number of bacteria in the leaves of transgenic plants reached to about 6× 105 cfu per mg, which is approximately 10% of that in the leaves of wild-type plants.
Discussion
Exogeneous application of MeJA causes alteration in a wide range of plant cellular metabolism, up- or down-regulating the expression of various genes. Most importantly, recently developed DNA microarray technology facilitates screening of a whole set of jasmonate-responsive genes.19-22) In parallel, MeJA-related responses against hormones, wounding, microbial attack, and abiotic stresses have been analyzed, making possible global analyses of expression profiles.23-26) Genes up-regulated by MeJA treatment include those for jasmonate biosynthesis, defense proteins, secondary Fig. 1. Northern blot analysis of transgenic Arabidopsis
overexpressing JMT. Total RNAs were isolated from rosette leaves of each four individual wild-type (WT) and transgenic (TR) Arabisdopsis. Ten micrograms of total RNA was electro- phoresed on 1.5% formaldehyde agarose gel and blotted onto nylon membranes. The blots were hybridized with each of the random primer-extended cDNA clones as indicated. Loading of equal amount of RNA was confirmed by staining the gel with ethidium bromide (data not shown).
Fig. 2. Dot blot analysis of defense gene expression. Four micrograms of total RNAs isolated from wild-type (WT) or JMT transgenic (TR) Arabidopsis were labeled with DIG-11- dUTP, and hybridized to a dot blot membrane at 42oC over- night. After post-hybridization washes, the membrane was blocked with 5% skim milk and treated with anti-DIG-POD- labeled antibody (1 : 5000). Hybridization signal on the mem- brane was detected by enhanced chemi-luminescence. DNA probes blotted on the membrane are PCR-amplified ESTs (Table 1) for the following proteins. 1, copper/zinc superoxide dismu- tase; 2, peoxidase; 3, 25S rDNA; 4, chlorophyll a/b-binding pro- tein; 5, thaumatin-like protein; 6, hevein-like protein; 7, basic chitinase; 8, β-1,3-glucanase; 9, extensin (Hrgp); 10, glu- tathione S-transferase; 11, thioredoxin.
Fig. 3. Disease resistance of the JMT transgenic Arabidopsis.
Seven-week-old Arabidopsis plants were infected with Pseudomonas syringae pv tomato DC3000 by spaying the bac- terial suspension (1× 108 cfu · ml−1). Infected rosette leaves were homogenized, and appropriate dilutions were plated onto King's medium B agar. After incubation at 28oC, the number of colonies on the medium was counted. Mean values and stan- dard deviations were calculated from six independent experi- ments with wild-type (open circles) and transgenic (closed circles) plants.
metabolism, cell wall formation, and stress-protective proteins. By contrast, genes involved in photosynthesis, such as ribulose bisphosphate carboxylase/oxygenase (Rubisco), chlorophyll a/b-binding protein, and light harvesting complex II, are down-regulated.
However, such chemically synthesized jasmonates used in most studies might have been contaminated by unnatural isomers. Thus, a clear distinction between the members of jasmonate family, including JA and MeJA, has not been made in each jasmonate-responsive cellular process. In addition, serious confusion has arisen occasionally due to unexpected results caused by nonphysiological concentration of the applied chemicals. In this regard, characterization of the transgenic plants transformed with the cellular component (JMT) that catalyzes the formation of endogenous MeJA has helped clarify the complexity of jasmonate-mediated plant responses.
Importance of JMT activation in the jasmonate-regulated responses was demonstrated in an experiment with transgenic Arabidopsis overproducing the gene.18) The transgenic plants contained elevated level of endogenous MeJA concentration and constitutively expressed various jasmonate-responsive genes in the absence of wounding or jasmonate treatment. By contrast, overexpression of a cytoplasm-localized flax AOS27) or the Arabidopsis AOS cDNA28) did not alter the basal level of jasmonates in transgenic tobacco and Arabidopsis, unless the tissues were damaged. AOS is known to be a key enzyme catalyzing the rate-limiting step of the jasmonate biosyntheis pathway.14,17) Moreover, transgenic potato plants constitutively expressing a plastidic flax AOS cDNA contained an increased level of JA, but transcription of jasmonate-responsive genes was not enhanced in these plants.29) Thus, MeJA formation has proven to be a critical control point for jasmonate-regulated plant metabolisms including defense responses. Indeed, the transgenic plant exhibited enhanced resistance to the virulent fungal pathogen Botrytis cinerea.18)
As observed in the present study, the transgenic plants contain high level of gene transcripts of PDF1.2 (Fig. 1). This gene encodes a toxic protein defensin, and is responsible for the defense against non-host fungal pathogens such as Botrytis cinerea, Aternaria brassicicola, and Pythium mastophorum.
In addition, expression of PR genes that encode β-1,3- glucanase (PR-2), basic chitinase (PR-3), hevein-like protein (PR-4), and thaumatin-like protein (PR-5) were significantly elevated in the transgenic plants (Fig 2). Among the genes showing increased level of transcript is the extensin gene Hrgp that is induced in response to wounding and pathogen attack to accumulate hydroxyproline-rich glycoproteins in the cell walls of plants.30,31) Oxidative stress-related genes for peroxidase, copper/zinc superoxide dismutase, and thioredoxin were also constitutively expressed in the transgenic plants. Rapid generation of active oxygen species including superoxide anion (O2−) and hydrogen peroxide (H2O2) has been described in many plant-pathogen interactions.32) Glutathione S-transferase gene was also up-
regulated, as it has been observed that expression of this gene is regulated in response to many forms of biotic and abiotic stresses.33)
The transgenic Arabidopsis overexpressing the JMT gene exhibited significantly fortified resistance against the infection with virulent pathogen Pseudomonas syringae pv tomato DC3000 (Fig. 3). This non-host virulent bacterial strain has constituted a model system for molecular genetic analysis of interactions with Arabidopsis Columbia ecotype.34) These results indicate that MeJA mediates plant defense responses against a broad spectrum of pathogens including fungi and bacteria.
As shown with the function of MeJA in defense gene activation, it is a remarkable feature of plant responses to produce and utilize volatile compound in cellular responses.35) In particular, MeJA has become a strong candidate for airborne signals that mediate interplant communication for defense responses.36) Further investigation of such gas-phase signaling mechanisms will permit an understanding of how plants use the volatiles to confront and cope with diverse and variable environments. Such advances will also provide a way toward manipulation of cellular signaling pathways to improve disease resistance of plants.
Acknowledgments. We thank Dr. Ingyu Hwang (Seoul National University) for providing the bacterial stain Pseudomonas syringae pv tomato DC3000. This work was supported by a grant from the Crop Functional Genomics Center (Korea) and in part by a grant from the ScigenHarvest Co., Korea. Financial supports including graduate research assistantships to Jung, Lyou, and Koo from the Brain Korea 21 Project of the Ministry of Education are also acknowledged.
References
1. Bowles, D. J. (1990) Defense-related proteins in higher plants. Annu. Rev. Biochem. 59, 873-907.
2. Hammond-Kosack, K. E. and Jones, J. D. G. (1996) Resis- tance gene-dependent plant defense responses. Plant Cell 8, 1773-1791.
3. Reymond, P. and Farmer, E. E. (1998) Jasmonate and sali- cylate as global signals for defense gene expression. Cur.
Opin. Plant Biol. 1, 404-411.
4. Dong, X. (1998) SA, JA, ethylene, and disease resistance in plants. Cur. Opin. Plant Biol. 1, 316-323.
5. Penninckx, I. A. M. A., Thomma, B. P. H. J., Buchala, A., Métraux, J.-P. and Broekert, W. F. (1998) Concomitant acti- vation of jasmonate and ethylene response pathway is required for induction of a plant defensin gene in Arabidop- sis. Plant Cell 10, 2103-2113.
6. Epple, P., Apel, K. and Bohlmann, H. (1995) An Arabidop- sis thaliana thionin gene is inducible via a signal transduc- tion pathway different from that for pathogenesis-related proteins. Plant Physiol. 109, 813-820.
7. Vigutelli, A., Wasternack, C., Apel, K. and Bohlmann, H.
(1998) Systemic and local induction of an Arabidopsis thio-
nin gene by wounding and patghogens. Plant J. 14, 285- 295.
8. Penninckx, I. A. M. A., Eggermont, K., Terras, F. R., Thomma, B. P. H. J., De Samblanx, G. W., Buchala, A., Métraux, J. P., Manners, J. M. and Broekaert, W. F. (1996) Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell 8, 2309-2323.
9. Thomma, B. P. H. J., Eggermont, K., Penninckx, I. A. M.
A., Mauch-Mani, B., Vogelsang, R., Cammue, B. P. A. and Broekaert, W. F. (1998) Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabi- dopsis are essential for resistance to distinct microbial pathogens. Proc. Natl. Acad. Sci. USA 95, 15107-15111.
10. Vijayan, P., Shockey, J., Lévesque, C. A., Cook, R. J. and Browse, J. (1998) A role for jasmonate in pathogen defense of Arabidopsis. Proc. Natl. Acad. Sci. USA 95, 7209-7214.
11. Pieterse, C. M. J., van Wees, S. C. M., van Pelt, J. A., Knoester, M., Laan, R., Gerrits, H., Weisbeek, P. J. and van Loon, L. C. (1998) A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell 10, 1571-1580.
12. Glazebrook, J. (1999) Genes controlling expression of defense responses in Arabidopsis. Cur. Opin. Plant Biol. 2, 280-286.
13. León, J., Rojo, E. and Sánchez-Serrano, J. J. (2001) Wound signalling in plants. J. Exp. Bot. 52, 1-9.
14. Creelman, R. A. and Mullet, J. E. (1997) Biosynthesis and action of jasmonates in plants. Annu. Rev. Plant Physiol.
Plant Mol. Biol. 48, 355-381.
15. Wasternack, C. and Hause, B. (2002) Jasmonates and octa- decanoids: signals in plant stress responses and develop- ment. Prog. Nucleic Acid Res. Mol. Biol. 72, 165-221.
16. Wasternack, C. and Parthier, B. (1997) Jasmonate-signalled plant gene expression. Trends Plant Sci. 2, 302-307.
17. Creelman, R. A. and Rao, M. V. (September 30, 2002) The oxylipin pathway in Arabidopsis. In The Arabidopsis book, Somerville C.R. and Meyerowitz E.M. (eds.) American Society of Plant Biologists, Rockville, MD, doi10.1199/
tab.0012 (http://www.aspb.org/publications/arabidopsis/).
18. Seo, H. S., Song, J. T., Cheong, J.-J., Lee, Y.-H., Lee, Y.- W., Hwang, I., Lee, J. S. and Choi, Y. D. 2001. Jasmonic acid carboxyl methyltransferase: A key enzyme for jas- monate-regulated plant responses. Proc. Natl. Acad. Sci.
USA 98, 4788-4793.
19. Reymond, P., Weber, H., Damond, M. and Farmer, E. E.
(2000) Differential gene expression in response to mechani- cal wounding and insect feeding in Arabidopsis. Plant Cell 12, 707-719.
20. Schenk, P. M., Kazan, K., Wilson, I., Anderson, J. P., Rich- mond, T., Somerville, S. C. and Manners, J. M. (2000) Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 97, 11655-11660.
21. Sasaki, Y., Asamizu, E., Shibata, D., Nakamura, Y., Kaneko, T., Awai, K., Amagai, M., Kuwata, C., Tsugane, T.,
Masuda, T., Shimada, H., Takamiya, K., Ohta, H. and Tabata, S. (2001) Monitoring of methyl jasmonate-respon- sive genes in Arabidopsis by cDNA macroarray: self-activa- tion of jasmonic acid biosynthesis and crosstalk with other phytohormone signaling pathways. DNA Res. 8, 153-161.
22. Ozturk, Z. N., Talamé, V., Deyholos, M., Michalowski, C.
B., Galbraith, D. W., Gozukirmizi, N., Tuberosa, R. and Bohnert, H. J. (2002) Monitoring large-scale changes in transcript abundance in drought- and salt-stressed barley.
Plant Mol. Biol. 48, 551-573.
23. Cheong, Y. H., Chang H.-S., Gupta, R., Wang, X., Zhu, T.
and Luan, S. (2002) Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiol. 129, 661-677.
24. Chen, W., Provart, N. J., Glazebrook, J., Katagiri, F., Chang, H. S., Eulgem, T., Mauch, F., Luan, S., Zou, G., Whitham, S. A., Budworth, P. R., Tao, Y., Xie, Z., Chen, X., Lam, S., Kreps, J. A., Harper, J. F., Si-Ammour, A., Mauch-Mani, B., Heinlein, M., Kobayashi, K., Hohn, T., Dangl, J. L., Wang, X. and Zhu, T. (2002) Expression profile matrix of Arabidopsis transcription factor genes suggests their puta- tive functions in response to environmental stresses. Plant Cell 14, 559-574
25. Kreps, J.A., Wu, Y., Chang, H.-S., Zhu, T., Wang, X. and Harper, J. F. (2002) Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol.
130, 2129-2141.
26. Lorenzo, O., Piqueras, R., Sánchez-Serrano J. J. and Sol- ano, R. (2003) ETHYLENE RESPONSE FACTOR1 inte- grates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15, 165-178.
27. Wang, C., Avdiushko, S., and Hidebrand, D. F. 1999. Over- expression of a cytoplasm-localized allene oxide synthase promotes the wound-induced accumulation of jasmonic acid in transgenic tobacco. Plant Mol. Biol. 40, 783-793.
28. Laudert, D., Schaller, F. and Weiler, E. W. (2000) Trans- genic Nicotiana tabacum and Arabidopsis thaliana plants overexpressing allene oxide synthase. Planta 211, 163-165.
29. Harms, K., Atzorn, R., Brash, A., Kühn, H., Wasternack, C., Willmitzer, L., and Peña-Cortés, H. 1995. Expression of a flax allene oxide synthase cDNA leads to increased endogenous jasmonic acid (JA) levels in transgenic potato plants but not to a corresponding activation of JA-respond- ing genes. Plant Cell 7, 1645-1654.
30. Showalter, A. M., Bell, J. N., Cramer, C. L., Bailey, J. A.
and Varner, J. E. (1985) Accumulation of hydroxyproline- rich glycoprotein mRNA in response to fungal elicitor and infection. Proc. Natl. Acad. Sci. USA 82, 6551-6555.
31. Mazau, D. and Esquerré-Tugayé, M. T. (1986) Hydroxypro- line-rich glycoprotein accumulation in the cell walls of plants infected by various pathogens. Plant Physiol. 80, 540-546.
32. Lamb, C. and Dixon, R. A. (1997) The oxidative burst in plant disease resistance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 251-275.
33. Marrs, K. A. (1996) The functions and regulation of glu- tathione S-transferases in plants. Annu. Rev. Plant Physiol.
Plant Mol. Biol. 47, 127-158.
34. Whalen, M. C., Innes, R. W., Bent, A. F. and Staskawicz, B. J. (1991) Plant Cell 3, 49-59.
35. Paré, P. W. and Tumlinson, J. H. 1999. Plant volatiles as a
defense against insect herbivores. Plant Physiol. 121, 325- 331.
36. Farmer, E. E. and Ryan, C. A. (1990) Interplant communi- cation: Air-borne methyl jasmonate induces synthesis of proteinase inhibitors in plant leaves. Proc. Natl. Acad. Sci.
USA 87, 7713-7716.