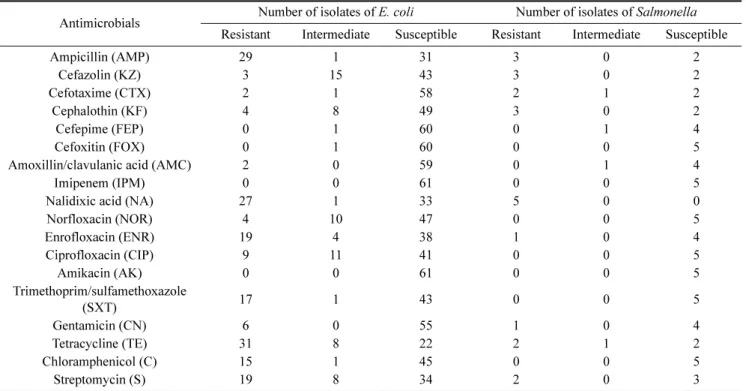
관련 문서
The purpose of this study is to define the concept of affordance in multi-media environment and to seek development plan of more efficient interface
mandibular P-18 inncisor from Nfic-deficient mice (×3,000)... In this study, histological and immunohistochemical studies were carried out to investigate
In the present study, consistent to that report, up-regulation of VEGF mRNA was observed in the colon cancer cell with the acquired resistance to 5-FU and this
In our search for the bioactive metabolites from myxobacteria, strains KM1001 and KM1041 of Sorangium cellulosum, were found to produce potent
Therefore, in order to find out more effective policy, it is essential to understand the technology commercialization in terms of the institutional support
In this study, it is shows that the stream of CO 2 is more effective in the reduction of unreacted TEGDMA and the increase of surface microhardness than that of N 2
Results: The human breast cancer cell subline MCF-7/MX5 cells selected in the presence of 5 µg/ml mitoxantrone (MX) were more resistant to MX (15.7... Western blot and
In this study, it is showed that female and blood type O are the significant factors associated with prolonged C/Epi CT value despite of aspirin discontinuation more