Journal of Bacteriology and Virology 2016. Vol. 46, No. 1 p.13 – 21 http://dx.doi.org/10.4167/jbv.2016.46.1.13
N-terminal Extension of Coat Protein of Turnip Yellow Mosaic Virus has Variable Effects on Replication, RNA Packaging, and Virion
Assembly Depending on the Inserted Sequence
Kwang-Hee Chae, Doyeong Kim and Tae-Ju Cho*
Department of Biochemistry, Chungbuk National University, Cheongju, Korea
Turnip yellow mosaic virus (TYMV) is a non-enveloped icosahedral virus composed of 20 kDa single coat proteins.
In this study, we modified the TYMV coat protein (CP) ORF by inserting an oligonucleotide linker corresponding to T7, HSV, Tat, (Arg)9, or (RxR)4 peptide at the 5'-end of the CP ORF and examined its effect on replication, RNA packaging, and virion assembly. The results showed that the constructs containing (Arg)9 and (RxR)4 sequences were barely capable of replication. The TYMV constructs containing T7 and Tat peptide produced virions that co-migrated with wild-type virions. However, the insertion of T7 and Tat sequences impaired genomic RNA (gRNA) accumulation and packaging, respectively. When only the CP gene was expressed, CPs with (Arg)9 or (RxR)4 successfully produced virus-like particles whose mobility was comparable to that of wild type. In the case of CP having a HSV tag, the virion band was not detected, although a sufficient amount of CP was produced. This indicates that CP with the HSV tag failed to assemble into virions.
Overall, the results suggest that TYMV replication, RNA packaging and virion assembly are strongly influenced by the insertion sequence.
Key Words: TYMV, CP modification, Replication, Virion assembly, RNA packaging
INTRODUCTION
Turnip yellow mosaic virus (TYMV), a positive strand RNA virus, is the type member of the genus Tymovirus and infects mainly Cruciferae plants. TYMV RNA contains three open reading frames (1). p206 and p69 are translated from the genomic RNA (gRNA) and function as replication protein and movement protein, respectively. Coat protein (CP) is not expressed from gRNA but from subgenomic RNA (sgRNA) (1). Virions are non-enveloped 28-nm T=3
icosahedrons composed of a single 20 kDa coat protein that is clustered in 20 hexameric and 12 pentameric subunits (2).
Plant viruses are an ideal drug delivery vector because they are highly symmetrical, robust, and biocompatible (3, 4).
Small drug molecules, including the anticancer drug doxo- rubicin, can be covalently attached to or encapsulated within virus particles (5). These viral particles can be delivered to specific tissues, including tumors, when labelled with specific peptides, e.g., Cowpea mosaic virus particles displaying peptide F56 specifically localized in tumors overexpressing vascular endothelial growth factor receptor 1 (6).
13
Received: October 27, 2015/ Revised: March 2, 2016/ Accepted: March 17, 2016
*Corresponding author: Tae-Ju Cho. Department of Biochemistry, Chungbuk National University, Cheongju, 28644, Korea.
Phone: +82-43-261-2309, Fax: +82-43-267-2306, e-mail: tjcho@chungbuk.ac.kr
**This study was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2012-0007588) and by a Chungbuk National University research grant, awarded in 2014.
○CCThis is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/license/by-nc/3.0/).
Original Article
Plant viruses are amenable to various chemical modifi- cations (5) and can be genetically modified. In genetic TYMV CP modifications, the CP N-terminus is flexible for modifications. Hayden et al. (7) replaced various TYMV CP parts and reported that modification in the N-terminal part was well tolerated. Replacement with the Plasmodium falciparum E71 epitope resulted in viable progeny. Recently, Powel et al. (8) showed that 5-26 N-terminal amino acids of TYMV CP were dispensable for virion assembly, although the removal of 26 amino acids decreased stability and in- creased porosity in virions. Virion assembly and RNA pack- aging properly occurred when TYMV CP was modified by inserting a c-Myc epitope peptide at the CP N-terminus (9).
Here, we further modified the CP genes to produce pro- teins with T7, HSV, Tat, (Arg)9, or (RxR)4 peptide at the CP N-terminus and examined the effect on viral replication, RNA packaging, and virion assembly. Tat, (Arg)9 and (RxR)4
peptides are known as cell penetrating peptides that enhance the entry of proteins, siRNAs, or other macromolecules into cells (10). The results obtained here show that TYMV repli- cation, RNA packaging, and virion assembly are strongly influenced by the sequence inserted at the 5'-end of CP ORF, in contrast with previous reports.
MATERIALS AND METHODS DNA constructs
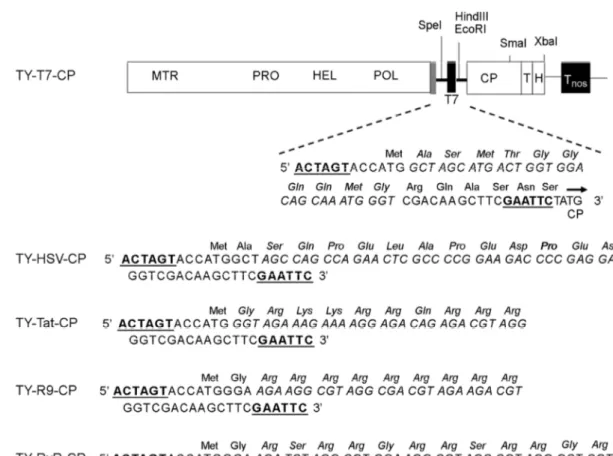
TY-HSV-CP, TY-T7-CP, TY-Tat-CP, TY-R9-CP and TY- RxR-CP constructs were made by inserting oligonucleotide linkers into the SpeI/EcoRI restriction sites of the TY-M- CP construct described previously (9). The nucleotide and amino acid sequences are shown in Fig. 1. To make CP constructs, the CP ORFs including the inserted sequence of the recombinant TYMV constructs were PCR-amplified with the following primers: TY(+)5596F (5'-CGCTGAG- TCTGAATTGCTTCACTACG-3') and TY(-)6223R (5'- GATCGAGAACTTAGGTGG-3'). The PCR product was digested with SpeI and XmaI, and was cloned into the pCB- M-CP construct, a derivative of the CP/pA construct. The pCB-M-CP and CP/pA constructs were described previously (9). The resulting constructs were designated as HSV-CP,
T7-CP, Tat-CP, R9-CP and RxR-CP.
Plant material
Agroinfiltration of the Agrobacterium tumefaciens har- boring the recombinant CP and TYMV constructs into Nicotiana benthamiana was carried out as described pre- viously (11). Seven days after agroinfiltration, the infiltrated leaves were collected. For RNA and protein extraction, the leaf samples were frozen in liquid nitrogen immediately after collection and were stored at -80℃. For encapsidation assay, the leaf sample was ground with 4 times its volume of phosphate buffer (pH 7.0). The homogenate was clarified by the addition of 0.2 volumes of chloroform, centrifuged briefly, and was stored at 4℃ until use.
Analysis of RNA
Ribonuclease protection assay for encapsidated RNA and Northern analysis were performed as previously described (11). Briefly, in the encapsidation assay, leaf extracts in sodium phosphate buffer were incubated with RNase A (5 μg/ml at the final concentration) before phenol extraction. In Northern blot hybridization, the membranes were hybridized with a DIG-labeled DNA probe representing the coat protein ORF (nt-5641 to nt-6231).
Analysis of coat protein
Coat protein (CP) was examined by 12.5% SDS- polyacrylamide gel electrophoresis (SDS-PAGE) followed by Western blot analysis. Leaf samples (0.1 g) were ground in 200 μl of 2X sample buffer and were boiled for 5 min. When leaf extracts and polyethyleneglycol (PEG)-precipitated samples were analyzed, two volumes of 2X sample buffer was added before boiling. Western blot detection used anti-TYMV rabbit antiserum in conjunction with HRP- conjugated goat anti-rabbit IgG (Bio-Rad, Hercules, CA, USA) and a chemiluminescent detection method using LuminataTM Forte (Millipore, Bedford, MA, USA).
Analysis of virions
Virions in leaf extracts or in PEG-precipitated samples were analyzed as described previously (9). PEG precipi-
tation of TYMV virions was carried out following the plant virus purification procedure described by Lane (12). The inoculated leaves were ground in the presence of three volumes of extraction buffer (1.3 M sodium acetate buffer, pH 4.5). The extract was mixed with 0.2 volume of chloro- form and the precipitant was removed by centrifugation.
0.5 volume of 30% PEG 8000 solution was then added to the supernatant, and the mixture was left at room tempera- ture for 1 hr. After centrifugation, the pellet was dissolved in storage buffer (60 mM sodium acetate buffer, pH 5.5 containing 1 mM EDTA and 1 mM MgCl2) and stored at 4℃. The samples containing virions were electrophoresed on a 1% agarose gel. For Northern analysis of virions, the
gel was treated with 0.2 N NaOH and then with 0.1 M Tris buffer (pH 7.5) before blotting to Hybond N+ membranes.
For Western analysis of virions, the agarose gel was treated with 0.1 M Tris buffer (pH 7.5), but not with NaOH, before blotting to Hybond ECL membranes.
RESULTS
Effect of N-terminal extension on TYMV replication and RNA packaging
TY-T7-CP, TY-HSV-CP, TY-Tat-CP, TY-R9-CP, and TY- RxR-CP constructs were prepared by replacing the c-Myc peptide sequence in the TY-M-CP construct (9) with the
Figure 1. TYMV constructs. TYMV CP was modified by inserting an oligonucleotide linker corresponding to the T7, HSV, Tat, (Arg)9, or (RxR)4 peptide into the SpeI/EcoRI sites (underlined) of TY-M-CP (9). The nucleotide and amino acid sequences of the peptides are indicated in italic. The methyltransferase (MTR), protease (PRO) helicase (HEL) and polymerase (POL) domains of TYMV genome are shown. T and H represent the tRNA-like structure and HDV (hepatitis delta virus) ribozyme, respectively. Tnos is the transcription terminator of the nos gene. The sgRNA promoter, tymobox, is represented by grey box.
sequence corresponding to T7, HSV, Tat, (Arg)9, or (RxR)4
peptide (Fig. 1). The inserted peptides were connected to the TYMV CP N-terminus via a linker comprising 6 or 7 amino acids. These modified TYMV constructs were intro- duced into Nicotiana benthamiana using the Agrobacterium- mediated T-DNA transfer system. Seven days after agro- infiltration, leaf extracts were prepared from the infiltrated leaves. Total and encapsidated RNA samples from agro- infiltrated leaves were examined by Northern blotting using the DIG-labeled DNA probe encoding CP (Fig. 2), as pre- viously described (11). The TY-M-CP, TY-T7-CP, TY-HSV- CP, TY-Tat-CP, TY-R9-CP, and TY-RxR-CP constructs are
hereafter abbreviated as TY-M, TY-T7, TY-HSV, TY-Tat, TY- R9, and TY-RxR, respectively. Total RNA analysis revealed that only TY-T7 replicated as efficiently as TY-M (Fig. 2A).
In other constructs, poor replication or an unusual pattern of viral RNA accumulation was observed. In TY-R9 and TY-RxR, TYMV RNA was not detected. Similarly, viral RNA was barely detected in TY-HSV. However, TYMV CP was detected in the TY-HSV sample when proteins were examined by anti-TYMV CP antibody (Fig. 2B). For con- firmation, Northern blotting was conducted again with six times more RNA sample in the TY-HSV case. The result showed the presence of viral RNAs (Fig. 2A). In TY-Tat Figure 2. Effect of the CP modification on replication and RNA packaging. (A and C) Northern analysis of total (A) and encapsidated (C) RNA. RNA samples were prepared from the N. benthamiana leaves collected 7 days after agroinfiltration. Viral RNAs were analyzed by Northern blot hybridization using the DIG-labeled probe representing the CP ORF. The positions of gRNA and sgRNA are indicated.
(B) CP expression analysis. Leaf extracts were examined by SDS-PAGE, then Western blotting using anti-TYMV CP antibody and HRP- conjugated secondary antibody. TYW represents a wild-type TYMV construct. TYΔCP2, TY-T7, TY-HSV, TY-Tat, TY-R9, and TY-RxR represent the TYΔCP2+SL2, TY-T7-CP, TY-HSV-CP, TY-Tat-CP, TY-R9-CP, and TY-RxR-CP constructs, respectively.
sample, only small gRNA amounts were detected, although a strong sgRNA band was observed.
Western blotting after SDS-polyacrylamide gel electro- phoresis (SDS-PAGE) of CP showed that coat proteins were produced from the TY-M, TY-T7, TY-HSV and TY-Tat con- structs (Fig. 2B). The samples used were leaf extracts. The construct TYΔCP2+SL2 (hereafter abbreviated TYΔCP2), where 32 amino acids (#141~#165 and #181~#187 amino acids) in the C-terminal part of CP are absent (13), was in- cluded as a negative control. The mutant CP from TYΔCP2 was detected by the anti-TYMV CP antibody. In the TY-R9 and TY-RxR construct cases, TYMV CPs were not detected.
Thus, TYMV replication was severely inhibited by the in- serted sequence corresponding to (Arg)9 or (RxR)4 peptide.
The virion (encapsidated) RNA analysis results are shown in Fig. 2C. The leaf extracts were treated with RNase A, followed by phenol extraction. Encapsidated and unencap- sidated viral RNAs would be protected and degraded, respectively, from RNase action. Compared with the total
RNA samples (Fig. 2A), the banding patterns were similar in TY-M and TY-Tat constructs. TY-T7 gRNA was barely observed in the "encapsidated RNA" sample, suggesting that it was not efficiently packaged. In TYΔCP2, encapsidated viral RNA bands were not evident, which was anticipated from the CP structural defect. In the TY-HSV case, the band shown at sgRNA position seems to be an artifact because no such band was observed in this sample in a separate experiment and no viral RNA band was detected in the mobility shift assay of virions (Fig. 3A).
Effect of the CP modification on virion assembly
To examine whether modified CPs influenced virion assembly, virions in leaf extracts were partially purified by polyethyleneglycol (PEG) precipitation and examined by electrophoresis on a 1.0% agarose gel, followed by Northern blotting of viral RNAs. PEG precipitation of TYMV virions was conducted following the TYMV purification described by Lane (12). Viral RNAs were liberated by alkali treatment Figure 3. Mobility shift assay of virions. (A) Examination of virion RNA by Northern analysis. PEG-precipitated virions were electro- phoresed on a 1% agarose gel and then treated with 0.2 N NaOH. The liberated virion RNA was blotted onto nylon membrane and examined by hybridization with a DIG-labeled CP DNA probe. (B) Examination of virions by Western analysis. After non-denaturing agarose gel electrophoresis, the virions were transferred to nitrocellulose membrane, and were probed with anti-TYMV CP. (C) Western analysis of CP after SDS-PAGE. CPs in PEG-precipitated samples (top panel) and in leaf extracts (bottom panel) were examined as described in Figure 2C.
of the gel after electrophoresis and blotted onto a nylon membrane, followed by hybridization using a DIG-labeled CP probe. Here one RNA band, unlike two (gRNA and sgRNA) bands in the conventional Northern blotting, is detected at the position of virions. The result showed that the TY-M, TY-Tat, and TY-T7 constructs produced the bands co-migrating with wild-type TYMV virions (Fig. 3A), suggesting that the modified CPs assembled correctly.
However, TYΔCP2, TY-HSV, TY-R9, and TY-RxR did not produce the wild-type virion band.
The absence of bands in TYΔCP2 and TY-HSV in Fig.
3A may represent failure in RNA packaging but not virion assembly. To directly test the virion assembly, virions were electrophoresed on an agarose gel and examined by Western blotting. Here, the virions in agarose gel were blotted onto a nitrocellulose membrane without prior treatment of the gel with NaOH solution. The virions were then probed with
anti-TYMV CP antibody (Fig. 3B). Again, the bands co- migrating with wild-type TYMV virions were observed in the TY-M, TY-Tat, and TY-T7 samples, while no such bands were detected with the TYΔCP2 and TY-HSV samples. The result indicated that the absence of the bands in TYΔCP2 and TY-HSV was due to aborted virion assembly.
Because the result in Fig. 3B contrasted with that of CP expression (Fig. 2B), we examined whether CP was pre- sent in the PEG-precipitated samples by SDS-PAGE and Western blotting. CP proteins were not detected in the PEG- precipitated samples of TYΔCP2 and TY-HSV-CP (top panel, Fig. 3C). When the leaf extracts were similarly examined, in contrast, CPs were present in TYΔCP2 and TY-HSV-CP samples (bottom panel, Fig. 3C), consistent with the result shown in Fig. 2B. This shows that CPs from TYΔCP2 and TY-HSV-CP constructs were present in leaf extract samples but not precipitated with PEG, possibly because the recom-
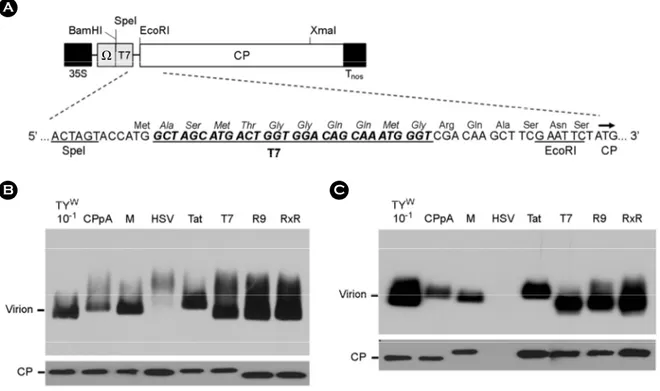
Figure 4. Assembly of the modified CPs expressed from CP constructs. (A) A CP construct expressing recombinant CPs. T7-CP is shown as a representative CP construct. Ω represents a translational enhancer from Tobacco mosaic virus. Virions in leaf extracts (B) or virions precipitated with PEG (C) were electrophoresed on a 1% agarose gel, and were examined by Western blot analysis (top panels), as described in Figure 3B. CPs in the samples were also examined by Western analysis after SDS-PAGE and blotting to nitrocellulose membrane (bottom panels). CP was detected as described in Figure 2C. CPpA, described previously (9), produces unmodified TYMV CPs.
Ω
binant CPs did not assemble into virions.
Because some inserted sequences appeared to interfere with TYMV replication, we prepared constructs containing only CP genes (Fig. 4A) to directly examine the modified CPs. To prepare CP constructs, CP ORFs including the inserted sequence in the recombinant TYMV constructs were PCR-amplified and cloned into the SpeI/XmaI sites of the pCB-M-CP construct (9). The recombinant CP genes were expressed under the influence of double CaMV 35S promoters, as observed for the TYMV constructs shown (Fig.
1). The CP mRNAs have a translation enhancer Ω derived from Tobacco mosaic virus and have a poly(A) tail at the 3'-ends. Except for the differences, CP would be the same as the proteins produced from the TYMV constructs described earlier.
Virion assembly with these modified CPs was examined by agarose gel electrophoresis followed by Western blotting (Fig. 4B and Fig. 4C). Here, all CPs including those from the R9-CP and RxR-CP constructs were successfully pro- duced. The CPpA construct produces unmodified TYMV CPs. Those CPs except for HSV-CP seemed to assemble into virus-like particles that were co-migrating with TYMV virions. In the CP case having HSV tag, a smear was observed around the position of wild-type virions (top panel, Fig. 4B) when the leaf extract was used. The HSV-CP protein was detected in the leaf extract by Western blotting after SDS-PAGE (bottom panel, Fig. 4B). When virions were precipitated with PEG, virion bands were detected in all modified CP samples except HSV-CP (top panel, Fig. 4C).
In these PEG-precipitated virion samples, CP was not detected by SDS-PAGE or Western blotting in the HSV-CP sample, consistent with the result in Fig. 3C. Thus, CP with the HSV tag failed to assemble into virions. The smear shown in the HSV-CP lane (top panel, Fig. 4B) may in- dicate that some assembly occurs with this HSV-modified CPs. However, the assembled particles, if any, are likely to be different from the authentic TYMV virions because they are excluded in the PEG precipitation step (Fig. 3C and Fig.
4C) and are unable to protect viral RNAs (Fig. 3A).
DISCUSSION
In TYMV replication, the 5'-untranslated region (UTR) and 3'-terminal sequence element, called an initiation box, are essential. Two 5'-UTR hairpins and a 17-nucleotide region upstream of the hairpins are crucial for efficient replication (14). In vitro studies using purified TYMV RNA- dependent RNA polymerase (RdRP) showed that template copying was predominantly controlled by the -CCA "initiation box" at the 3'-end, which needs to be non-base-paired and readily accessible (15, 16). 3'-terminal tRNA-like structure (TLS) is also important for TYMV replication. TYMV TLS shows the characteristic features of authentic tRNAs. For example, eEF1A binds to TLS, enhancing translation (1).
However, the tRNA conformation should be disrupted for replication. Recently, Colussi et al. (17) reported that G4-C76 base pairs in the TLS region serve as a linchpin to trigger TYMV TLS adoption of two differently folded states. The delicate conformation switch of TLS may have been in- terfered with by the R9, RxR and HSV sequence insertions.
Alternatively, the insertion may have disturbed the secondary structure of the 3'-terminal region of TYMV gRNA, leading to poor recognition of the 3'-terminal initiation box by TYMV RdRP.
(Arg)9, or (RxR)4 peptides are rich in arginine residues.
Such basic N-terminal sequence of CP, called arginine-rich RNA-binding motif (ARM), has been implicated in genome packaging in some spherical plant RNA viruses (18). The ARM, however, is not present in TYMV CP, and the role contacting and neutralizing viral RNA is served by poly- amines that are co-encapsidated with viral RNA (1). Insertion of arginine-rich peptides, thus, might instead have disturbed TYMV RNA packaging, leading to poor accumulation of viral RNA. However, the results obtained with TY-R9 or TY-RxR are not readily explained by the possible negative effect on RNA packaging, since virtually no viral RNA was detected in TY-R9 or TY-RxR, whereas some viral RNA accumulation was observed in the TYΔCP2 showing no RNA packaging (Fig. 2). It was also reported that about 10% of viral RNA, compared to wild type, accumulated in
the leaf cells transfected with a TYMV mutant RNA unable to produce CP (1). Therefore, it is highly likely that the in- sertion of (Arg)9, or (RxR)4 sequence has a direct detrimental effect on replication rather than on RNA packaging.
The case of TY-Tat seems more complicated; gRNA accumulation was impaired but with a less conspicuous effect on sgRNA. This could be due to aborted synthesis of (-)-strand. When the (-)-strand synthesis is incomplete, the resultant RNA would lack the replication element at the 5'-end, leading to decreased production of (+)-gRNA. sgRNA synthesis would not be affected because the sgRNA pro- moter, tymobox, resides upstream of CP ORF (19) and is far downstream of the 5'-element.
The situation with TY-T7 shows that gRNA packaging is inefficient, while sgRNA packaging is unaffected. This could be because the extra T7 peptide modifies the TYMV capsid structure and excludes larger RNA packaging. A crystal structure study of the TYMV virions revealed that the N-terminal part of some TYMV CPs (A subunit) is flexible and move inside/outside the virions through the opening at the pentamer apex (2). If this mobility was affected by the T7 peptide such that the T7 peptide stayed inside the virion, the space inside the virion could be restricted/modified and the packaging of bigger RNAs be excluded. In some plant RNA viruses, including the Cucumber mosaic virus, the virion structure is flexible and accommodates RNA twice as large as that found in wild type (18). Recently, we observed that the efficiency of TYMV gRNA encapsidation sharply decreased when an extra sequence of ≥2.2 kb was inserted (20). Considering the size constraint in TYMV RNA pack- aging, the change in virion structure caused by the T7 peptide, if any, could have resulted in gRNA exclusion.
REFERENCES
1) Dreher TW. Turnip yellow mosaic virus: transfer RNA mimicry, chloroplasts and a C-rich genome. Mol Plant Pathol 2004;5:367-75.
2) Canady MA, Larson SB, Day J, McPherson A. Crystal structure of turnip yellow mosaic virus. Nat Struct Biol 1996:3:771-81.
3) Steinmetz NF, Evans DJ. Utilization of plant viruses in biotechnology. Org Biomol Chem 2007;5:2891-902.
4) Steinmetz NF. Viral nanoparticles as platforms for next- generation therapeutics and imaging devices. Nano- medicine 2010;6:634-41.
5) Yildiz I, Shukla S, Steinmetz NF. Applications of viral nanoparticles in medicine. Curr Op Biotechnol 2011;
22:901-8.
6) Brundel FM, Lewis JD, Desitto G, Steinmetz NF, Manchester M, Stuhlmann H, et al. Hydrazone ligation strategy to assemble multifunctional viral nanoparticles for cell imaging and tumor targeting. Nano Lett 2010;
10:1093-7.
7) Hayden CM, Mackenzie AM, Skotnicki ML, Gibbs A.
Turnip yellow mosaic virus isolates with experimentally produced recombinant virion proteins. J Gen Virol 1998;
79:395-403.
8) Powel JD, Barbar E, Dreher TW. Turnip yellow mosaic virus forms infectious particles without the native beta- annulus structure and flexible coat protein N-terminus.
Virology 2012;422:165-73.
9) Shin H-I, Chae K-H, Cho T-J. Modification of Turnip yellow mosaic virus coat protein and its effect on virion assembly. BMB reports 2013;46:495-500.
10) Verdurmen WPR, Brock R. Biological responses towards cationic peptides and drug carriers. Trends Pharmacol Sci 2011;32:116-24.
11) Cho T-J, Dreher TW. Encapsidation of genomic but not subgenomic Turnip yellow mosaic virus RNA by coat protein provided in trans. Virology 2006;356:126-35.
12) Lane LC. Propagation and purification of RNA plant viruses. Methods Enzymol 1986;118:687-9.
13) Shin H-I, Cho T-J. Read-through mutation in the coat protein ORF suppresses Turnip yellow mosaic virus subgenomic RNA accumulation. J Bacteriol Virol 2013;
43:54-63.
14) Shin H-I, Tzanetakis IE, Dreher TW, Cho T-J. The 5'-UTR of Turnip yellow mosaic virus does not include a critical encapsidation signal. Virology 2009;387:427 -35.
15) Deiman BALM, Verlaan PWG, Pleij CWA. In vitro transcription by the turnip yellow mosaic virus RNA
polymerase: a comparison with the alfalfa mosaic virus and brome mosaic virus replicases. J Virol 2000;74:
264-71.
16) Singh RN, Dreher TW. Specific site selection in RNA resulting from a combination of nonspecific secondary structure and -CCR- boxes: initiation of minus strand synthesis by turnip yellow mosaic virus RNA-dependent RNA polymerase. RNA 1998;4:1083-95.
17) Colussi TM, Costantino DA, Hammond JA, Ruehle GM, Nix JC, Kieft JS. The structural basis of transfer RNA mimicry and conformational plasticity by a viral RNA.
Nature 2014;511:366-70.
18) Rao ALN. Genome packaging by spherical plant RNA viruses. Annu Rev Phytopathol 2006;44:61-87.
19) Schirawski J, Voyatzakis A, Zaccomer B, Bernardi F, Haenni AL. Identification and functional analysis of the turnip yellow mosaic tymovirus subgenomic promoter.
J Virol 2000;74:11073-80.
20) Kim H-B, Chae K-H, Cho T-J. Genome size constraint in replication and packaging of Turnip yellow mosaic virus. J Bacteriol Virol 2014;44:188-96.