Fundamentals of
Molecular Biology
4 Fundamentals of Molecular Biology
Chapter Outline
Heredity, Genes, and DNA
Expression of Genetic Information
Recombinant DNA
Detection of Nucleic Acids and
Proteins
Introduction
The focus of molecular biology concerns the mechanisms responsible for
transmission and expression of genetic information.
Fundamental experiments on model organisms such as bacteria and viruses have provided information on molecular mechanisms that operate in all organisms.
Recombinant DNA technology allowed fundamental principles and experimental approaches to be extended to eukaryotic cells.
Recombinant DNA allowed isolation and characterization of individual genes, and determination of complete genome sequences.
All organisms inherit genetic information specifying their structure and function from their parents.
All cells arise from pre-existing cells, so the genetic material must be
replicated and passed from parent to progeny at each cell division.--- 왜
Heredity, Genes, and DNA
The classical principles of genetics were deduced by Gregor
Mendel in 1865, from his experiments with pea plants.
He studied well-defined traits such as seed color, and could predict patterns of inheritance by assuming that each trait is determined by a pair of inherited factors, now called genes. One gene copy (allele) specifying each trait is inherited from
each parent.
Example: strains with identical alleles specifying yellow (Y) or green (y) seeds, are crossed.
The progeny (F1 generation) are hybrids (Yy) and have yellow seeds: yellow is dominant, green is recessive.
Heredity, Genes, and DNA
Genotype is the genetic makeup of an individual. Phenotype is the resulting physical appearance.
The genotype of the F1 generation: Yy The phenotype: yellow.
Mendel’s results were ignored until 1900 when their importance was recognized.
Shortly afterward, chromosomes were identified as the carriers of genes.
Q) Differences between prokaryotic and eukaryotic genes?
Heredity, Genes, and DNA
Most cells of plants and animals are diploid: they have two copies of each chromosome.
Formation of germ cells (sperm and egg) involves a type of cell division (meiosis) in which only one
member of each chromosome pair is transmitted to the progeny cell.
The sperm and egg are haploid: they have only one copy of each chromosome.
At fertilization, two haploid cells are combined to make a new diploid cell.
Heredity, Genes, and DNA
Fundamentals of mutation, genetic linkage, and relationships between genes and chromosomes were established by
experiments with the fruit fly Drosophila melanogaster. In the early 1900s, mutations (genetic alterations) were
observed in Drosophila that affected characters such as eye color and wing shape.
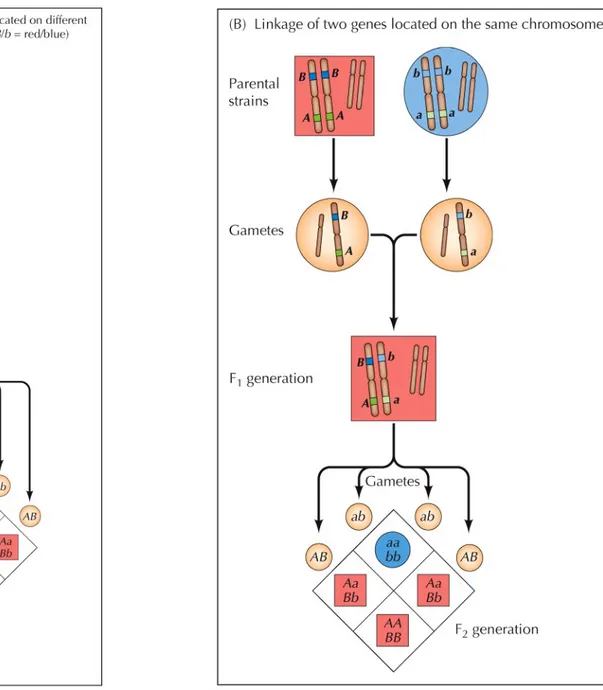
Genetic linkage
Breeding experiments showed that some of these genes are inherited independently of each other.
This suggested that the genes are on different chromosomes that segregate independently during meiosis.
Genes located on the same chromosome are inherited together and are said to be linked.
Heredity, Genes, and DNA
The relationship between genes and proteins began to emerge in 1909:
It was realized that the inherited disease
phenylketonuria results from a genetic defect in the enzyme needed to metabolize the amino acid phenylalanine -- 발달장애 , 경련 : hydroxylase 결핍 In 1941, experiments with the fungus Neurospora
crassa by Beadle and Tatum found more evidence linking genes with the synthesis of enzymes.
Mutant strains of Neurospora required particular amino acids for growth.
Heredity, Genes, and DNA
Each mutation resulted in a deficiency in a specific metabolic pathway, which is governed by enzymes. This led to the conclusion that genes specify enzyme
structure. : one gene and one protein
one gene and one enzyme
It is now known that some genes specify RNAs rather than proteins.
Heredity, Genes, and DNA
Evidence that DNA is the genetic material first came from experiments with the bacterium that causes pneumonia (Pneumococcus).
A “transforming principle” was responsible for
inducing the genetic transformation of one strain of the bacteria to another.
The transforming principle was later identified as DNA when it was shown that the activity of the transforming principle is abolished by enzymatic digestion of DNA: but not by digestion of proteins.
Heredity, Genes, and DNA
Other studies with viruses confirmed that DNA was the genetic material.
It was shown that when a bacterial virus infects a cell, the viral DNA must enter the cell in order for the virus to replicate (not the viral protein).
Heredity, Genes, and DNA
The structure of DNA was determined in 1953 by Watson and Crick, using information from X-ray crystallography and ideas on hydrogen bonding in the α helix of proteins.
DNA is a double helix with the sugar- phosphate backbones on the outside of the molecule.
The bases are on the inside; hydrogen bonds are formed between purines and pyrimidines on
opposite chains.----DNA breathing
The base pairing is very specific: A always pairs with T and G with C.
Heredity, Genes, and DNA
DNA replication
Complementary base pairing suggested the mechanism for DNA replication.
The DNA molecule separates, and each strand
becomes a template for synthesis of a new strand:
Semiconservative replication: one strand of
parental DNA is conserved in each progeny DNA molecule.
Heredity, Genes, and DNA
Experimental support for semi-conservative DNA
replication:
Meselson and Stahl grew E. coli in medium labeled with the heavy isotope 15N.
The heavier DNA could be separated from light DNA (with 14N) by equilibrium ultracentrifugation in a CsCl
solution.
E. coli grown with 15N were transferred to 14N medium
and allowed to replicate once more.
The DNA from these bacteria was intermediate in weight.
Heredity, Genes, and DNA
Further evidence: an enzyme purified from E. coli
(DNA polymerase) could catalyze DNA replication
in vitro.
In the presence of DNA to act as a template, DNA
polymerase directed the incorporation of nucleotides into a complementary DNA molecule.
Genes determine the structure of proteins.
The information in DNA is specified by the order of the four bases.
Expression of Genetic Information
Proteins are made up of 20 different amino acids, the sequence of which determines the protein function.
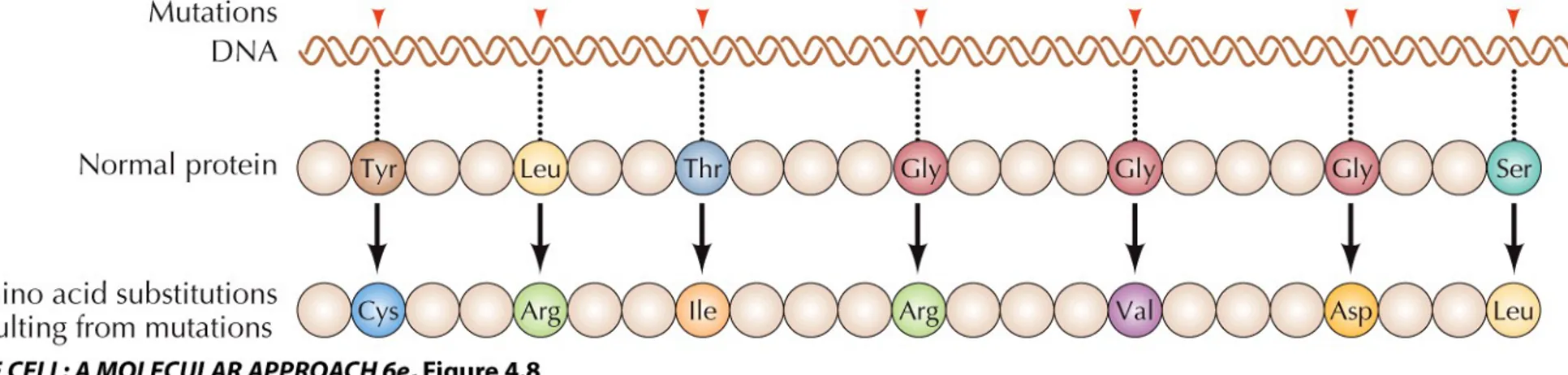
The first direct link between a genetic mutation and alteration in a protein was made in 1957.
Patients with inherited sickle-cell anemia had
hemoglobin that differed from normal by a single amino acid substitution.
The next step was determining the relationship between DNA and proteins.
Expression of Genetic Information
The simplest explanation: the order of nucleotides in DNA specifies the order of amino acids in proteins (colinearity).
Mutations (changes in the nucleotide sequence) should lead to corresponding changes in the protein.
Using E. coli, Yanofsky and colleagues mapped a series of mutations in the gene that encodes an enzyme for tryptophan synthesis.
The relative positions of the amino acid alterations were the same as those of the corresponding
Expression of Genetic Information
Although DNA appeared to specify the order of amino acids in proteins, it did not necessarily follow that DNA itself directs protein synthesis. DNA is located in the nucleus of eukaryotic cells,
Expression of Genetic Information
RNA was the likely candidate to be an intermediate because of its similar structure.
RNA is single-stranded, its sugar is ribose, and it contains uracil (U) instead of thymine (T), but this does not affect base-pairing.
The pathway for the flow of genetic information: DNA → RNA → Protein
is now known as the central dogma of molecular biology.
Expression of Genetic Information
RNA is synthesized from DNA templates (transcription); proteins are synthesized from RNA templates (translation).
RNA polymerase catalyzes synthesis of messenger RNA (mRNA) from a DNA template.
Ribosomal RNA (rRNA) is a component of ribosomes, sites of protein synthesis.
Transfer RNAs (tRNAs) serve as adaptor molecules that align amino acids along the mRNA template.
Each amino acid is attached by a specific enzyme to its appropriate tRNA.
Base pairing between the tRNA and a complementary sequence on the mRNA directs the attached amino acid to its correct position on the mRNA template.
Expression of Genetic Information
How can four nucleotide bases specify the sequence of 20 amino acids?
The nucleotides are used as triplets to encode the different amino acids: genetic code.
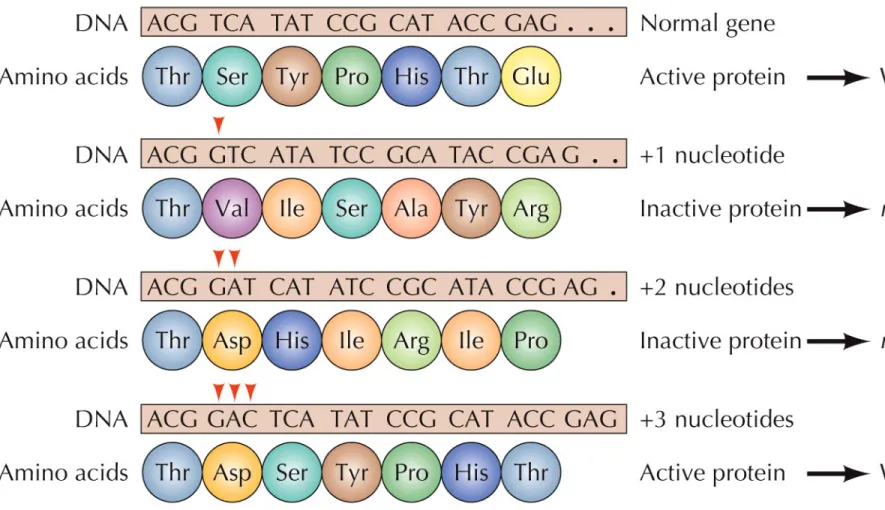
Evidence for the triplet code came from bacteriophage T4 with mutations in a gene called rII.
Mutants with the addition of one or two bases always exhibited the mutant phenotype.
Changes in three bases frequently led to the wild-type phenotype.
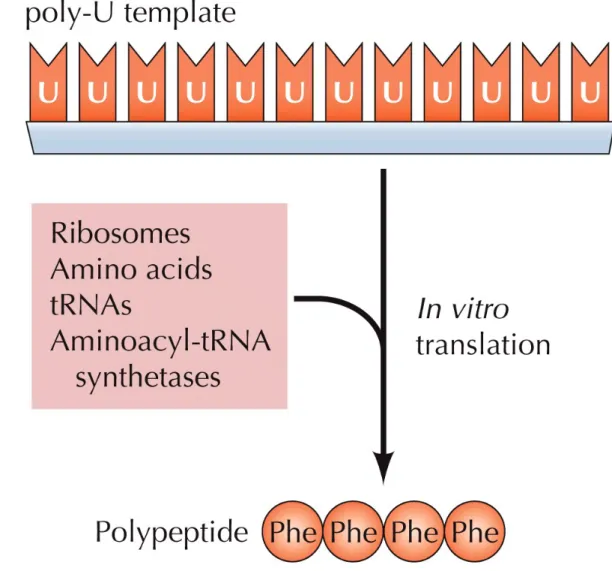
Figure 4.12 The triplet UUU encodes phenylalanine
Assigning nucleotide triplets to their
corresponding amino acids was done using
in vitro systems that
could carry out protein synthesis (in vitro
translation).
The systems contain ribosomes, amino acids, tRNAs,
enzymes, and synthetic mRNA with known
Expression of Genetic Information
All 64 possible triplets (called codons) were assigned in this way.
61 specify an amino acid; three are stop codons
that signal the termination of protein synthesis. The code is degenerate: many amino acids are
specified by more than one codon.
With few exceptions, all organisms utilize the same genetic code—strong support for the conclusion that all present-day cells evolved from a common ancestor.
Expression of Genetic Information
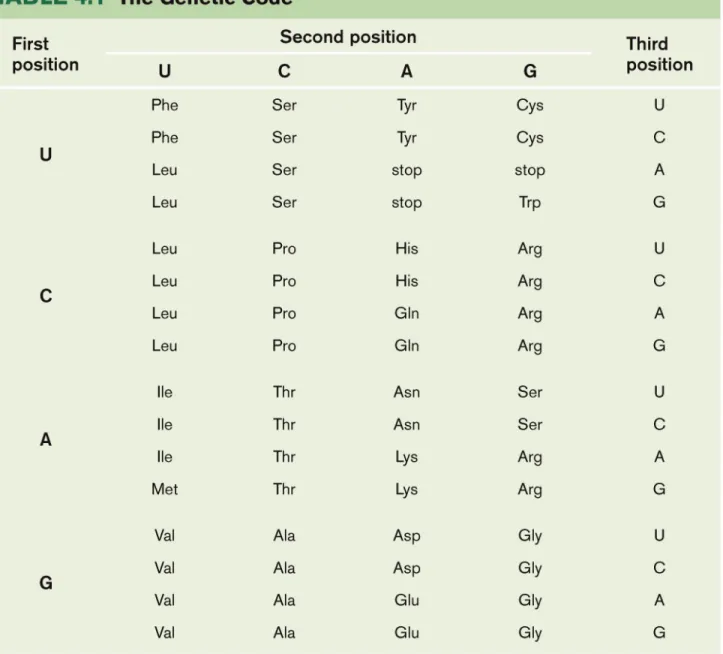
Some viruses contain RNA instead of DNA.
The mode of replication of viral RNA was determined by studies of RNA bacteriophages of E. coli.
These viruses encode an enzyme that catalyzes
synthesis of RNA from an RNA template (
RNA-directed RNA synthesis).
Most animal viruses replicate in this way, but one group (RNA tumor viruses) requires DNA synthesis in
infected cells.
These viruses (now called retroviruses) replicate via synthesis of a DNA intermediate, a DNA provirus.
Expression of Genetic Information
This hypothesis was initially met with
disbelief because it reverses the
central dogma.
Later, an enzyme that catalyzes
synthesis of DNA from an RNA
template (reverse transcription) was
discovered.
Key Experiment, Ch. 4, p. 117
Howard M. Temin
Nobel Prize
in Physiology or Medicine 1975
Expression of Genetic Information
Reverse transcription has other broad implications. It also occurs in cells and is frequently responsible
for transposition of DNA from one chromosomal location to another.
Reverse transcriptase can be used experimentally to generate DNA copies of any RNA molecule.
This has allowed mRNAs of eukaryotic cells to be studied using the molecular approaches currently applied to the manipulation of DNA.
Recombinant DNA
Recombinant DNA technology allows scientists to isolate, sequence, and manipulate individual genes from any type of cell.
It has enabled detailed molecular studies of the structure and function of eukaryotic genes and genomes, and revolutionized our understanding of cell biology.
Restriction endonucleases: enzymes that cleave DNA at
specific sequences.
First identified in bacteria, where they provide defense against the entry of foreign DNA.
Bacteria have a variety of restriction endonucleases that cleave DNA at more than 100 distinct recognition sites.
Recombinant DNA
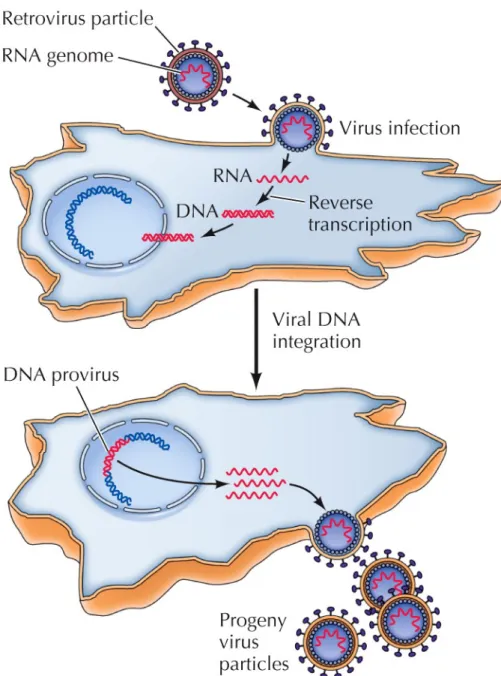
EcoRI recognizes the sequence GAATTC.
This sequence is present at five sites in DNA of
bacteriophage λ, so it is digested into six fragments ranging from 3.6 to 21.2 kb long.
1 kilobase (kb) = 1000 base pairs
The fragments can be separated by gel
electrophoresis:
A gel of agarose or polyacrylamide is placed between two electrodes and the sample is added to the gel. Nucleic acids are negatively charged so they migrate toward the positive electrode.
Recombinant DNA
Smaller molecules move through the gel more
rapidly, allowing the fragments to be separated by size.
The order of restriction fragments can also be determined, and maps of restriction sites
generated.
Detailed restriction maps of viral DNA molecules have been produced.---RFLP
Recombinant DNA
For larger DNA molecules, restriction
endonuclease digestion alone does not provide sufficient resolution.
For example, the human genome would yield more than 500,000 EcoRI fragments.
Purified DNA fragments can be obtained through molecular cloning.
In molecular cloning a DNA fragment is inserted into a DNA molecule (a vector) that can replicate independently in a host cell.
Recombinant DNA
The result is a recombinant molecule or molecular
clone.
Fragments of human DNA can be cloned in plasmid
vectors: small circular DNA molecules that can replicate independently in bacteria.
Recombinant plasmids with human DNA inserts can be introduced into E. coli, where they replicate along with the bacteria to yield millions of copies of plasmid DNA. The fragment can be isolated from the rest of the plasmid
DNA by restriction endonuclease digestion and gel electrophoresis, allowing a pure fragment of human DNA to be analyzed and further manipulated.
Recombinant DNA
Restriction endonucleases cleave the
recognition sequences at staggered
sites, leaving overhanging
single-stranded tails that can associate with
each other by complementary base
pairing.
DNA ligase can then seal the ends
Recombinant DNA
Synthetic DNA “linkers” containing desired restriction endonuclease sites can be added to the ends of any DNA fragment.
This allows virtually any fragment of DNA to be ligated to a vector and isolated as a molecular clone.
RNA can also be cloned.
RNA is copied using reverse transcriptase. The resulting DNA (cDNA) is ligated to a vector DNA.
This allows mRNA to be isolated as a molecular clone, and allows exploration of the noncoding sequences in eukaryotic genes (introns).
Recombinant DNA
Plasmids are often used for cloning DNA inserts up to a few thousand base pairs long.
Plasmids have an origin of replication—the DNA sequence that signals the host DNA polymerase to start replication.
Plasmid vectors also carry genes that confer resistance to antibiotics, so bacteria carrying the plasmids can be selected for.--- Selection markers
Recombinant DNA
Bacteriophage λ vectors can accommodate larger
DNA fragments.
Sequences not needed for virus replication are
removed and replaced with unique restriction sites for insertion of cloned DNA.
The recombinant molecules are then put into E. coli, where they replicate to yield millions of progeny
Recombinant DNA
For even larger fragments of DNA, five major types of vectors are used.
1. Cosmid vectors contain bacteriophage λ
sequences, origins of replication, and genes for antibiotic resistance, so they are able to replicate as plasmids in bacterial cells.
2. Bacteriophage P1 vectors allow recombinant molecules to be packaged in vitro into P1 phage particles and replicated as plasmids in E. coli.
Recombinant DNA
3. P1 artificial chromosome (PAC) vectors also have bacteriophage P1 sequences, but are
introduced directly as plasmids into E. coli.
4. Bacterial artificial chromosome (BAC) vectors are derived from a naturally occurring plasmid of
E. coli (the F factor).
5. Yeast artificial chromosome (YAC) vectors contain yeast origins of replication and other
sequences that allow them to replicate as linear chromosome-like molecules in yeast
Recombinant DNA
Nucleotide sequencing aids the study of protein
structure, gene sequences that regulate
expression, and gene function.
DNA sequencing is usually done with automated
systems.
One method is based on premature termination
of DNA synthesis.
Recombinant DNA
Dideoxynucleotides are included along with the
normal nucleotides. They are labeled with different fluorescent dyes.
The dideoxynucleotides stop DNA synthesis because no 3 OH group is available for addition of the next nucleotide.
The fragments are separated by gel electrophoresis, and a laser beam excites the fluorescent dyes.
Recombinant DNA
Next-generation sequencing
allows DNA
to be sequenced faster and less
expensively.
These methods simultaneously determine
sequences of millions of templates by
Recombinant DNA
Molecular cloning is also used to make large amounts of protein for study.
Many proteins in cells are present in small amounts and can’t be purified.
Cloned genes can be used to engineer vectors that lead to high levels of gene expression in bacteria or
eukaryotic cells.
In bacteria, cDNA is cloned into a plasmid or phage vector (an expression vector).
Inserted genes can be expressed at levels as high as 10% of the total bacterial protein.
Recombinant DNA
Expression in eukaryotic cells instead of bacteria
may be needed, (e.g., if
posttranslational
modification
of the protein is required).
Cloned genes are inserted into virus vectors.
One such system uses infection of insect cells by
Detection of Nucleic Acids and Proteins
Detection of specific nucleic acids and
proteins is important for a variety of studies:
Mapping of genes to chromosomes
Analysis of gene expression
Localization of proteins to subcellular
organelles.
Detection of Nucleic Acids and Proteins
To isolate large amounts of a single DNA molecule, an
alternative to molecular cloning is the polymerase chain
reaction (PCR), developed in 1988.
DNA polymerase is used in vitro for repeated replication of a defined segment of DNA.
A specific region of DNA can be amplified if the nucleotide sequence surrounding the region is known.
Two primers are designed to initiate DNA synthesis in opposite directions at the desired point.
The synthetic primers are 15–20 bases long.
The reaction starts by heating the template DNA to 95°C to separate the strands.
Detection of Nucleic Acids and Proteins
The DNA polymerase used (Taq polymerase) is a heat-stable enzyme from Thermus aquaticus, a bacterium that lives in hot springs.
The temperature is then lowered to allow primers to pair and DNA synthesis proceeds.
Two DNA molecules are synthesized from one template.
The cycle can be repeated multiple times, with a two-fold increase in DNA for each cycle.
Detection of Nucleic Acids and Proteins
PCR amplification can be performed rapidly and automatically.
RNA can also be amplified by PCR if reverse
transcriptase is used to synthesize a cDNA copy first.
If enough of a gene sequence is known so that
primers can be made, PCR can selectively amplify it from complex mixtures, such as total cell DNA or
RNA.
This extraordinary sensitivity has made PCR an important method for a variety of applications.
Detection of Nucleic Acids and Proteins
The key to detection of specific nucleic acid sequences is base pairing.
In nucleic acid hybridization, DNA strands are separated by high temperatures; when cooled, they re-form double-stranded molecules by
complementary base pairing.
Cloned DNA can be labeled with radioactive
nucleotides or nucleotides modified to fluoresce. This labeled DNA is then used as a probe that
hybridizes with complementary DNA or RNA in complex mixtures.
Detection of Nucleic Acids and Proteins
Southern blotting:
DNA is digested with a restriction
endonuclease, and the fragments separated
by gel electrophoresis.
The gel is then overlaid with a nitrocellulose
or nylon membrane to which the DNA
fragments are transferred (blotted). The
Detection of Nucleic Acids and Proteins
Northern blotting is used for detection
of RNA instead of DNA.
It is often used in studies of gene
expression, for example, to determine
whether specific mRNAs are present.
Detection of Nucleic Acids and Proteins
Recombinant DNA libraries are
collections of clones that contain all the
genomic or mRNA sequences of a
particular cell type.
A genomic library of human DNA can be
made by cloning random DNA fragments
of about 15 kb in a bacteriophage λ
Detection of Nucleic Acids and Proteins
Any gene for which a probe is available
can be isolated from a recombinant
library.
cDNA clones can be used as probes to
isolate corresponding genomic clones.
Or a gene cloned from one species (e.g.,
mouse) can be used to isolate a related
gene from a different species (e.g.,
Detection of Nucleic Acids and Proteins
Hybridization to DNA microarrays allows tens of
thousands of genes to be analyzed simultaneously. A DNA microarray is a glass slide or membrane filter
onto which oligonucleotides or fragments of cDNAs are printed by a robotic system in small spots at a high density.
One application of DNA microarrays is in studies of gene expression.
Example: a comparison of the genes expressed by two different types of cells.
Detection of Nucleic Acids and Proteins
In situ hybridization can be used to detect
homologous DNA or RNA sequences in cell extracts, chromosomes, or intact cells.
Hybridization of fluorescent probes to specific cells or subcellular structures is analyzed by microscopic examination.
Detection of Nucleic Acids and Proteins
Antibodies can be used as protein probes.
Antibodies are proteins produced by immune system cells (B lymphocytes) that react against foreign
molecules (antigens).
Different antibodies recognize unique antigens. Antibodies can be generated by inoculation of an
animal with any foreign protein.
Monoclonal antibodies can be produced by
culturing clonal lines of B lymphocytes from immunized animals (usually mice).
Detection of Nucleic Acids and Proteins
Two common methods using antibodies: 1. Immunoblotting (Western blotting)
Proteins are separated by size by
SDS-polyacrylamide gel electrophoresis (SDS-PAGE).
The negatively charged detergent sodium dodecyl sulfate (SDS) denatures the protein and gives it an overall negative charge.
The proteins will migrate towards the positive electrode and are then reacted with labeled antibodies.
Detection of Nucleic Acids and Proteins
2. Immunoprecipitation:
Cells are incubated with radioactive amino acids to label their proteins, then incubated with antibodies. The resulting antigen-antibody complexes are
isolated and subjected to electrophoresis.
Antibodies can be used to visualize proteins in intact cells.
Cells can be stained with antibodies labeled with fluorescent dyes or tags visible by electron
Gene Function in Eukaryotes
In classical genetics, gene function has been revealed by the altered phenotypes of mutant organisms.
It is now possible to study the function of a cloned
gene directly by reintroducing it into eukaryotic cells. Yeasts (e.g., Saccharomyces cerevisiae) are used in
studies of eukaryotic cells because they are easily grown in culture, reproduce rapidly, and have a
small genome.
Mutants that have specific nutrient requirements can be easily isolated.
Gene Function in Eukaryotes
Temperature-sensitive mutants encode
proteins that are functional at one
temperature (permissive temperature) but
not another (nonpermissive temperature).
The ability to isolate temperature-sensitive
mutants has allowed identification of
genes controlling many fundamental cell
processes.
Gene Function in Eukaryotes
A gene corresponding to any yeast mutation
can be cloned based on its functional
activity.
Yeast genes encoding a wide variety of
essential proteins have been identified in
this manner.
In many cases, such genes have also been
useful in identifying and cloning related
Gene Function in Eukaryotes
Cloned DNA can also be introduced into
plant and animal cells (gene transfer).
Transfection uses infectious viral
Gene Function in Eukaryotes
Other methods include:
•
Direct microinjection
into the nucleus
•
Coprecipitation of DNA with calcium
phosphate to form small particles that are
taken up by the cells
•
Incorporation of DNA into
liposomes
that
fuse with the plasma membrane
•
Exposure of cells to brief electric pulses
that open pores in the plasma membrane
(
electroporation
)
Gene Function in Eukaryotes
In most of the cells, the DNA is transported to the nucleus, and is transcribed for several days:
transient expression.
In about 1% of cells, the foreign DNA is integrated into the genome and transferred to progeny cells at cell division.
If the transfected DNA contains a selectable marker, the stably transformed cells can be isolated and studied.
Gene Function in Eukaryotes
Animal viruses, especially retroviruses,
can be used as vectors to introduce
cloned DNAs into cells.
Gene Function in Eukaryotes
Cloned genes can also be introduced
into the germ line of multicellular
organisms.
Mice that carry foreign genes
(transgenic mice) are produced by
microinjection of cloned DNA into the
pronucleus of a fertilized egg.
Gene Function in Eukaryotes
Embryonic stem (ES) cells are also
used to get cloned genes into mice.
Cloned DNA is introduced into ES cells
in culture, then transformed cells are
introduced back into mouse embryos.
The offspring are chimeric: a mixture of
cells that arise from normal and
transfected embryonic cells.
Gene Function in Eukaryotes
Introducing specific mutations into cloned DNAs (in
vitro mutagenesis) is a powerful tool to study
expression and function of eukaryotic genes.
Sometimes called reverse genetics: a mutation is introduced into a gene first, and its functional
consequence is determined second.
In vitro mutagenesis allows detailed characterization of
the functional roles of both regulatory and protein-coding sequences of cloned genes.
The most common method uses synthetic
oligonucleotides to generate changes in a DNA sequence.
Gene Function in Eukaryotes
To determine the role of a cloned gene,
activity of the normal gene copy must
be eliminated.
Homologous recombination: the
mutated copy of the cloned gene
replaces the normal gene copy in the
chromosomal DNA.
This occurs frequently in yeast but is
rare in mammalian cells.
Gene Function in Eukaryotes: transgenic vs knockout mice
Genes can be inactivated in mouse embryonic stem cells, which can grow into transgenic mice.
The mice yield progeny with mutated copies of the gene on both homologous chromosomes.
The effects of inactivation of a gene can then be investigated in the context of the intact animal.
Homologous recombination has been used to systematically inactivate (knockout) every gene in yeast.
A collection of genome-wide yeast mutants is available for scientists to use to study the function of any desired gene. In mice, there is an international effort to knockout all mice
genes to develop a genome-wide collection of mutant mice for research.
Gene Function in Eukaryotes
Other approaches interfere with gene
expression or function.
Antisense nucleic acids
are RNA or
single-stranded DNA complementary to
the mRNA of the gene of interest
(antisense)---cf)
siRNA vs miRNA
.
They hybridize with the mRNA and block its
translation into protein.
Gene Function in Eukaryotes
RNA interference (RNAi)
was first discovered
in C. elegans.
Fire and Mello
found that injection of
double-stranded RNA
inhibited expression of a gene
with a complementary mRNA sequence.
Double-stranded RNA resulted in extensive
degradation of the target mRNA, whereas
single-stranded antisense RNA had only a
minimal effect.
Gene Function in Eukaryotes
When double-stranded RNAs are introduced into cells, they are cleaved into short interfering RNAs (siRNAs) by an enzyme called Dicer.
The siRNAs associate with a complex of proteins known as the RNA-induced silencing complex (RISC), where mRNA is cleaved.
The discovery of RNA interference demonstrated a role for double-stranded RNAs in gene regulation. This has been developed into a powerful
experimental tool for inhibiting expression of target genes.
Key Experiment, Ch. 4, p. 144 (1) Andrew Z. Fire Stanford Univ Craig C. Mello Univ of Massachusetts Nobel Prize
Gene Function in Eukaryotes