저작자표시-비영리-변경금지 2.0 대한민국 이용자는 아래의 조건을 따르는 경우에 한하여 자유롭게 l 이 저작물을 복제, 배포, 전송, 전시, 공연 및 방송할 수 있습니다. 다음과 같은 조건을 따라야 합니다: l 귀하는, 이 저작물의 재이용이나 배포의 경우, 이 저작물에 적용된 이용허락조건 을 명확하게 나타내어야 합니다. l 저작권자로부터 별도의 허가를 받으면 이러한 조건들은 적용되지 않습니다. 저작권법에 따른 이용자의 권리는 위의 내용에 의하여 영향을 받지 않습니다. 이것은 이용허락규약(Legal Code)을 이해하기 쉽게 요약한 것입니다. Disclaimer 저작자표시. 귀하는 원저작자를 표시하여야 합니다. 비영리. 귀하는 이 저작물을 영리 목적으로 이용할 수 없습니다. 변경금지. 귀하는 이 저작물을 개작, 변형 또는 가공할 수 없습니다.
Regulation of mRNA Expression Through 3’UTR
Sequences of IL6 Gene in Amino Acid-deprived
HeLa Cells.
by
Jung Hee Kang
Major in Molecular Medicine
Department of Biomedical Sciences
The Graduate School, Ajou University
Regulation of mRNA Expression Through 3’UTR
Sequences of IL6 Gene in Amino Acid-deprived
HeLa Cells.
by
Jung Hee Kang
A Dissertation Submitted to The Graduate School of
Ajou university in Partial Fulfillment of the Requirements for
The Degree of Master of Biomedical Sciences
Supervised by
Yong-Joon Chwae, M.D., Ph.D.
Major in Molecular Medicine
Department of Biomedical Sciences
The Graduate School, Ajou University
This certifies that the dissertation
of Jung Hee Kang is approved.
SUPERVISORY COMMITTEE
최 용 준
Yong-Joon Chwae
박 선
Sun Park
권 명 희
Myung-Hee Kwon
The Gradute School, Ajou University
i -ABSTRACT –
Regulation of mRNA Expression Through 3’UTR
Sequences of IL6 Gene in Amino Acid-deprived
HeLa Cells.
ER stress responses and autophagic responses triggered by amino acid-deprivation of cancer cells activate STAT3 and NF-κB transcriptional factors subsequently to induce IL6 cytokines. Here, it was investigated whether 3’UTR of IL6 mRNA played a role in IL6 induction through cellular stress responses. In the studies with GFP-reporter containing 3’UTR of IL6 mRNA, it was elucidated that 3’UTR contributed to increase of IL6 mRNA after amino acid-starvation, and its motifs seemed to be broadly distributed according to deletion mutants studies of IL6 3’UTR. NF-κB functioned in 3’UTR to partially contribute to stabilization of IL6 mRNA according to knock-down experiments of p65, NF-κB subunit. Finally, treatments of p38 MAP kinase inhibitor, SB203580 markedly blocked mRNA stabilization through 3’UTR as well as IL6 induction after amino acid-starvation, suggesting that p38-MK2/3-TTP pathway is evidently activated and implicated in stabilization of IL6 mRNA during starvation of cancer cells. Conclusively, these studies indicate that stress signals via NF-κB and p38 lead to mRNA stabilization through 3’UTR sequences, and their molecular mechanism would need to be further studied.
Key words : IL6 cytokines, AU rich element, 3’untranslation region, post-transcription, mRNA stability and translation
ii
TABLE OF CONTENTS
ABSTRACT ··· ⅰ TABLE OF CONTENTS ··· ⅱ LIST OF FIGURES ··· ⅳ LIST OF TABLES ··· ⅴ ABBREVIATION ··· ⅴ Ⅰ. INTRODUCTION ··· 1Ⅱ. MATERIALS AND METHODS ··· 3
A. Cell, antibodies and other reagents··· 3
B. Expression constructs and lentiviral transfections ··· 3
C. Real time RT PCR ··· 4
D. Western blot ··· 4
Ⅲ. RESULTS ··· 6
A. IL6 induction in amino acid-deprived HeLa cell ··· 6
B. Constructions of GFP-reporter plasmids containing 3’UTR of IL6 mRNA ··· 6
C. Effects of 3’UTR on induction of IL6 reporter in amino acids-deprived HeLa cells ·· 8
D. Effect of partial 3’UTR on induction of IL6 reporter in amino acids-deprived HeLa cells ··· 8
E.GFP expressions in HeLa cells infected with pCMV or pCMV-UTR before/after amino acid-deprivation ··· 8
iii
F. Effects of STAT3 and NF-kB transcriptional factors on IL6 3’UTR in IL6 induction of
amino acid-deprived HeLa cells ··· 14
G. Effects of MAP kinase inhibitor on IL6 3’ UTR in amino acids-deprived HeLa cells ··· 14
Ⅳ. DISCUSSION ··· 19
Ⅴ. CONCLUSION ··· 21
REFERENCES ··· 22
iv
LIST OF FIGURES
Fig. 1. IL6 induction in amino acid-deprived HeLa cells. ··· 7
Fig. 2. Schematic diagram of GFP-reporter plasmids containing IL6 promoter, CMV promoter, and/or 3’UTR sequences of IL6 genes ··· 9
Fig. 3. Complete or partial 3’UTR sequences of IL6 genes included in lentiviral GFP reporter plasmids. ··· 10
Fig. 4. Effects of 3’UTR on induction of IL6 reporter in amino acids-deprived HeLa cells. ··· 11
Fig. 5. Effects of partial 3’UTR on induction of IL6 reporter in amino acids-deprived HeLa cells ··· 12
Fig. 6. GFP expressions in HeLa cells infected with pCMV or pCMV-UTR before/after amino acid-deprivation. ··· 13 Fig. 7. Effects of STAT3 and NF-κB transcriptional factor on IL6 3’UTR in IL6 induction of
amino acid-deprived HeLa cells. ··· 16 Fig. 8. Effects of Oncostatin M and TNF α on IL6 3’UTR in HeLa cells. ··· 17 Fig. 9. Effects of MAP kinase inhibitor on IL6 3’UTR in amino acids-deprived HeLa cells. · 18
v
LIST OF TABLE
Table 1. Primers for Cloning ··· 5 Table 2. Primers for Real Time PCR ··· 5
1
Ⅰ. INTRODUCTION
IL-6 is a cytokine with pleomorphic biologic effects expressed in fibroblasts, monocytes/macrophages, endothelium, keratinocytes, and other mesenchymal and epithelial cells in response to a variety of noxious stimuli, including tumor necrosisfactor-α (TNFα), IL-1β, LPS, platelet-derived growth factor, and interferons(Sehgal, 1990). It is also produced constitutively in some lymphomas, sarcomas, and carcinomas and is elevated in patients with systemic bacterial infections(Akira &Kishimoto, 1992; Lust, 1994). In addition, IL6 can be induced intrinsically without external stimuli, especially in cellular stressful conditions such as hypoxia (Ali et al, 1999; Yamauchi-Takihara et al, 1995) and ER stress (Shi et al, 2009; Yoon et al, 2011).
IL-6 gene and their interacting partners may vary depending on the cell type as well as the extracellular stimuli. Functional cis-regulatory elements described to date in the human IL-6 promoter include IRF-1 (Faggioli et al, 1997), AP-1 (Dendorfer et al, 1994), C/EBPβ(Akira &Kishimoto, 1997; Isshiki et al, 1990), Sp1 (Kang et al, 1996) and NF-κB (Libermann& Baltimore, 1990)binding sites.
One of integral methods in controlling gene expression is by post-transcriptional mechanisms that regulate mRNA stability and protein translation. This type of regulation is definitely important as microarray analysis has detected that 40–50% of the changes in inducible gene expression occur at the level of mRNA stability(Cheadle et al, 2005). Regulatory elements of mRNA that play a critical role in post-transcriptional regulation typically reside within the 3’untranslated region (3’UTR) of the mRNA(Garneau et al, 2007). MicroRNAs (miRNAs) are small non-coding RNAs that have important roles in a post-transcriptional gene regulation through imperfect base pairing to the 3’UTR of its target mRNA in order to control mRNA stability and translation (Fabian et al, 2010). Currently, it is estimated that nearly 1,000 miRNAs function in humans and have be enpredicted to regulate approximately 60% of all protein coding genes (Friedman et al, 2009). Another fundamental mechanism of 3’UTR-mediated post-transcriptional regulation is through association with various RNA-binding proteins that select target mRNAs containing
2
adenylate- and uridylate (AU)-rich elements (AREs) within their 3’UTR (Garneau et al, 2007). These proteins also guide target sequences for miRNA-mediated regulation, implying the possibility for crosstalk between miRNA and ARE binding proteins (von Roretz&Gallouzi, 2008).
One of the hallmarks of cancer cells is to be resistant and survive from cellular stressful conditions. This is obtained by stress responses of cancers that confer evasion mechanisms from cell death on tumor cells. In this context, it is recently known that amino acid-deprivation of cancer cells triggers autophagy and ER stress responses to activate NF-κB and STAT3 transcriptional factors, resulting in induction of IL6 cytokines. In the present work, it was investigated what is the role of 3’ UTR of IL6 mRNA and its affecting factors during IL6 induction in amino acids-deprivation of cancer cells.
3
Ⅱ. MATERIALS AND METHODS
A. Cell, antibodies and other reagents.
HeLa cells, human cervical cancer cell line, were obtained from the American Type Culture Collection (CCL-2) and cultured in minimal essential media (MEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 2 mM Lglutamine, 100 U/ml penicillin and 100 μg/ml streptomycin. Anti-Cop GFP( Cat.#AB501) antibody were purchased from evrogen, NF-kB p65(A)(SC-109) antibody purchased from Santa Cruz biotechnology, and anti-STAT3 (9139) antibody were purchased from Cell Signaling. TNF-α and Oncostatin M(295-OM-0505) cytokine were purchased from R&D systems. Inhibitor reagents, p38 inhibitor-SB203580 (Cell Signaling #5633 ) and MEK1 inhibitor-PD98059 (Cell Signaling #9900) were purchased from Cell Signaling. JNK inhibitor-SP600125 (SC-200635) purchased from Cell Signaling ,
B. Expression constructs and lentiviral transfections.
All the constructs were subcloned into pCDH-EF2-MCS-T2A-Puro, a lentiviral vector for cDNA expression (System Biosciences, CD520A-1). Partial IL6 promoter and 5'UTR sequences (-568 to +70) were PCR-amplified from HeLa genomic DNA, cloned into pCDH-GFP (pCDH-IL6Pro), in which SpeI/SalI fragments of pCDH-EF2-MCS-T2A-Puro were ligated with PCR-amplified GFP2 sequences. And Cloned into pIL6 pro-UTR which SalI fragments pCDH-IL6Pro were ligated PCR-amplified 3¢ untranslated regions(3¢UTR). AUUUA motife site overlapped six. Partial CMV promoter were PCR-amplified from pcDNA3.1 vector, cloned into pCMV-GFP (pCDH-CMV Pro), in which SpeI/SalI fragments of pCDH-EF2-MCS-T2A-Puro were ligated with PCR-amplified GFP2 sequences. And Cloned into pCMV pro-UTR which SalI fragments pCMV-GFP were ligated PCR-amplified 3¢ untranslated regions(3¢UTR). AUUUA motife site overlapped six. pCMV-UTR d1 has overlapped AUUUA motife site four and pCMV-pCMV-UTR d2 has overlapped AUUUA motife site three.
The fragments length is IL6 promoter - 845bp, GFP- 660bp, CMV promoter- 571bp, 3'UTR- 428bp, 3'UTR d1 -336bp, 3'UTR d2 -220bp. The sequences of primers are described
4
in Table 1. All the lentiviral vectors were transfected to 293TN cells (SystemBiosciences, LV900A-1) with Lipofectamine 2000 transfection reagents (Invitrogen, 11668019). Particles were collected 2days after the transfection of lentiviral plasmids, and infected into HeLa cells.
C. Real-time reverse transcription (RT)-PCR
Cells were harvested at the indicated time points after treatment. Total RNAs were isolated by using an RNeasy kit (Qiagen, 74104). PrimeScript® RT reagent Kit (TaKaRa, RR037A) was used to reverse transcribe mRNA into cDNA. PCR was then performed on an ABI PRISM 7000
machine (Applied Biosystems) using SYBR Premix Ex Taq II(TaKaRa, RR041A). The sequences of primers are described in Table 2. Analysis of each sample was performed twice for each experiment and data in the figures are reported as relative quantification (RQ): average values of 2-ΔΔCT ± SD.
D. Western blot analysis.
At the indicated time points, cells were lysed in high salt lysis buffer [20 mM Tris-HCl (pH 8.0), 1%Triton X-100, 2 mM EDTA, 1 mM phenylmethylsulfonyl fluoride], incubated on ice for 20 min and microcentrifuged for 20 min to remove cell debris. Twenty μg of total cell lysate were subjected to SDS-polyacrylamide electrophoresis. The proteins were then electrotransferred to polyvinylidene difluoride membrane and incubated overnight with antibodies at 4°C. Subsequently, the membranes were incubated with peroxidase-conjugated secondary antibodies (Pierce, 31430 and 31460) for 1 hr at room temperature and the signal was detected using an enhanced chemiluminescence(ECL) detection kit (Amersham Biosciences, RPN2209).
5 Table 1. Primers for Cloning
6
Ⅲ. RESULTS
A. IL6 induction in amino acid-deprived HeLa cells.
It is known that amino acid deprivation of cancer cells induces IL6 mRNA and its secreted protein, which is dependent on STAT3 and NF-κB transcriptional factors. Jak2/STAT3 pathway is activated by Nox-generated reactive oxygen species (ROS), in which early autophagic processes would be implicated(Yoon et al, 2010). NF-κB is shown to be activated by IKK-independent mechanisms related to ER stress signals such as ROS, cytosolic calcium, and translational attenuation of IκBα mRNA compared to p65. Very interestingly, STAT3 and NF-κB cooperatively work in proximal IL6 promoter as an identical nuclear complex to induce IL6 (Yoon et al, 2011). For confirming the previous findings, responses of HeLa cells against nutritional defects were observed on IL6 mRNA induction. As shown in figure 1, HeLa cells incubated in minimal essential media (Salt, Gluc, AA, Vit), Hank’s salt solution (Salt), Hank’s salt solution with essential amino acids (Salt, AA), Hank’s salt solution with essential vitamins (Salt, Vit), Hank’s salt solution with essential amino acids and vitamins (Salt, AA, Vit), Hank’s salt solution with glucose and essential amino acids (salt, Glc, AA) showed a little or no IL6 induction, however, the cells incubated in Hank’s salt solution with glucose, so called Hank’s balanced salt solution (HBSS) (Salt, Glc), and Hank’s salt solution with glucose and essential vitamins showed markedly increased IL6 mRNA, implying that amino acids-deprivation in the presence of glucose transduced the signals for IL6 induction, consistent with the previous observations (Yoon et al, 2010; Yoon et al, 2011).
B. Constructions of GFP-reporter plasmids containing 3’UTR of IL6 mRNA.
Expression of specific genes are firstly guided by regulation of promoter activity governed by transcriptional complexes including transcriptional factors, additionally recent studies have been showing that post-transcriptional control of cytokine mRNA is also very important factors for regulation of gene expressions, which is mainly controlled 3’untranslated regions (3’UTR) (Anderson et al, 2004). Therefore, in the present work, roles of 3’UTR in IL6 induction of amino acid-deprived cancer cells were investigated. At first, for accomplishing these purposes, several constructs of GFP-reporters including IL6 3’UTR were made. As
7
Fig. 1. IL6 induction in amino acid-deprived HeLa cells.
HeLa cells were incubated in minimal essential media (MEM) (Salt, Gluc, AA, Vit), Hank’s salt solution (Salt), Hank’s balanced salt solution (HBSS) (Salt, Glc), Hank’s salt solution with amino acids (Salt, AA), Hank’s salt solution with vitamins (Salt, Vit), Hank’s salt solution with amino acids and vitamins (Salt, AA, Vit), HBSS with amino acids (Salt, Glc,
AA), HBSS with vitamins (Salt, Glc, Vit) for 15 hrs. IL6 mRNA levels were determined by
8
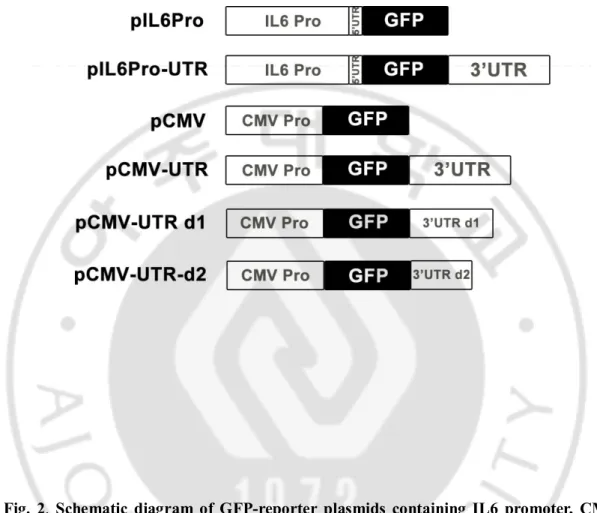
shown in figure 2, GFP reporters containing partial IL6 promoter sequences with or without whole IL6 3’UTR were made (pIL6Pro and pIL6Pro-UTR), and in GFP reporters containing CMV promoter (pCMV), partial or complete IL6 3’UTR sequences were added (pCMV-UTR, pCMV-UTR d1, pCMV-UTR d2). Partial or complete IL6 3’UTR sequences used for construction of GFP-reporters were described in figure 3.
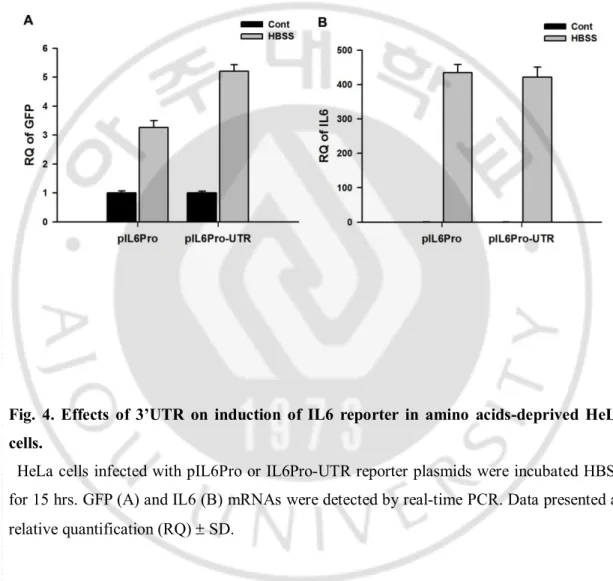
C. Effects of 3’UTR on induction of IL6 reporter in amino acids-deprived HeLa cells. For examining the effect of 3’UTR in IL6 mRNA induction after starvation, reporter activities in GFP-reporter with IL6 promoter were compared with those in GFP-reporter with IL6 promoter and 3’UTR. HeLa cells infected with the GFP-reporters containing IL6 promoter and 3’UTR showed more GFP mRNA induction compared to cells with IL6 promoter only after amino acid-deprivation (Fig. 4A), although there was no difference of IL6 mRNA induction (Fig. 4B). It suggests that signaling mechanisms that increased IL6 mRNA through 3’UTR can be working during amino acid-starvation of cancer cells.
D. Effects of partial 3’UTR on induction of IL6 reporter in amino acids-deprived HeLa cells.
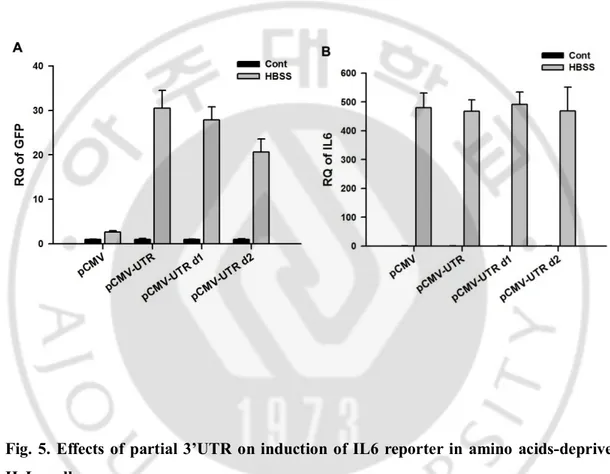
For searching the regions of IL6 3’UTR associated with starvation-induced IL6 mRNA induction, reporter plasmids with partially deleted forms of 3’UTR were made; pCMV-UTR d1 contains partial IL6 3’UTR deleted in distal 93 nucleotides: pCMV-UTR d2 contains partial IL6 3’UTR deleted in distal 208 nucleotides (Fig. 3). Partial deletion of 3’UTR in GFP-reporters showed decreased reporter activities compared to full 3’UTR, but not completely blocked reporter activities, even though no difference in IL6 induction in all the cells (Fig. 5 A and B), implying that the motifs within 3’UTR implicated in IL6 mRNA stability are broadly located extending full 3’UTR of IL6 mRNA.
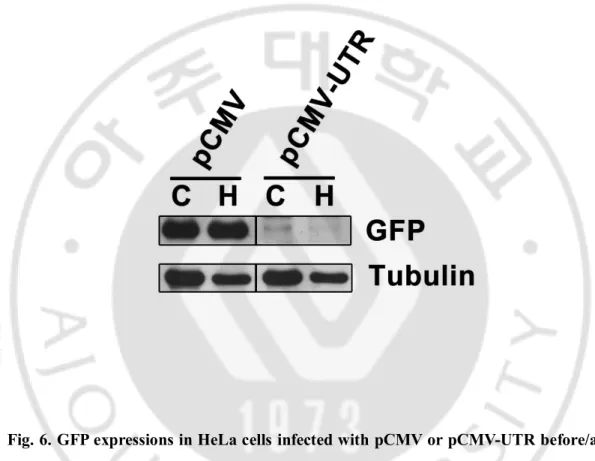
E. GFP expressions in HeLa cells infected with pCMV or pCMV-UTR before/after amino acid-deprivation
GFP protein expression in HeLa cells with pCMV-UTR plasmid were detected by western blotting and compared with that in cells with pCMV plasmid. As shown in figure 6, GFP
9
Fig. 2. Schematic diagram of GFP-reporter plasmids containing IL6 promoter, CMV promoter, and/or 3’ UTR sequences of IL6 genes.
Partial promoter regions and 5’UTR of IL6 genes were clone into pGF1 lentiviral plasmids with or without 3’ UTR sequences of IL6 genes (pIL6 Pro and pIL6 Pro-UTR). CMV promoters were cloned into GF1 with or without complete 3’UTR sequences, partial 3’ UTR sequnces of IL6 genes (pCMV, pCMV-UTR, pCMV-UTR d1, pCMV-UTR d2).
10
Fig. 3. Complete or partial 3’ UTR sequences of IL6 genes included in lentiviral GFP reporter plasmids.
Complete or partial IL6 3’UTR sequences (IL6 3’ UTR, IL6 3’UTR d1, IL6 3’UTR d2) were PCR-amplified from HeLa cDNA and cloned into pGF1 lentiviral plasmids. Boxed sequences indicate AU-rich elements (ARE) in the IL6 3’ UTR regions.
11
Fig. 4. Effects of 3’UTR on induction of IL6 reporter in amino acids-deprived HeLa cells.
HeLa cells infected with pIL6Pro or IL6Pro-UTR reporter plasmids were incubated HBSS for 15 hrs. GFP (A) and IL6 (B) mRNAs were detected by real-time PCR. Data presented as relative quantification (RQ) ± SD.
12
Fig. 5. Effects of partial 3’UTR on induction of IL6 reporter in amino acids-deprived HeLa cells.
HeLa cells infected with pCMV, pCMV-UTR, pCMV-UTR d1, or pCMV-UTR d2 reporter plasmids were incubated HBSS for 15 hrs. GFP (A) and IL6 (B) mRNAs were detected by real-time PCR. Data presented as relative quantification (RQ) ± SD.
13
Fig. 6. GFP expressions in HeLa cells infected with pCMV or pCMV-UTR before/after amino acid-deprivation.
HeLa cells infected with pCMV or pCMV-UTR plasmids were incubated in HBSS (H) or complete media (C) for 15 hrs. expressions of GFP and α tubulin were measured by Western blots.
14
expression was observed in the cells expressing pCMV control plasmid but no difference, regardless of nutritional statuses, and the cells expressing pCMV-UTR showed little GFP protein before and after amino acid-starvation. It is possible that 3’UTR of IL6 mRNA also have repressive signals of protein translation that cause to hardly detect signals.
F. Effects of STAT3 and NF-κB transcriptional factors on IL6 3’UTR in IL6 induction of amino acid-deprived HeLa cells.
IL6 expression in starved cancer cells are totally dependent on promoter activation of STAT3 and NF-κB nuclear complexes(Yoon et al, 2011), hence for elucidating the possibility that STAT3 and NF-κB also function in IL6 3’UTR and subsequently mRNA stabilization, HeLa cells knocked-down in STAT3 and NF-κB p65 subunit were established and decreases of STAT3 and p65 were confirmed by Western blot (Fig. 7A). Then, GFP expression from the reporter with pCMV-UTR was measured by real-time PCR before and after amino acid-starvation. As shown in figure 7B, STAT3 knock-down showed no influence on GFP reporter activities compared with non-targeting shRNA but p65 knock-down showed partially decreased reporter activities. Therefore, NF-κB signals appears to partially contribute to stabilization of IL6 mRNA through 3’UTR, and further studies about these points would be needed. Moreover, supporting these, oncostatin M treatment known to activate Jak2/STAT3 pathway and TNFα, well-known activator of canonical NF-κB pathway (Tanaka &Miyajima, 2003; Xiao & Fu, 2011)were no effect on IL6 3’UTR although TNFα increase IL6 mRNA to comparable levels with amino acid-deprivation (Fig. 8 A and B). Finally, knock-downs of STAT3 and p65 showed decreased IL6 mRNA induction compared to NT control shRNA consistent with previous observations (Yoon et al, 2010; Yoon et al, 2011) (Fig. 7C).
G. Effects of MAP kinase inhibitor on IL6 3’UTR in amino acids-deprived HeLa cells. 3’ UTR of IL6 mRNA has been known to have AU-rich elements (ARE), which is controlled by various RNA-binding factors functioning mainly negatively but also positively (Espel, 2005) as indicated in figure 3. Recent findings are showing that ARE sequences of TNFα can be positively controlled by p38 MAP kinase-MK2/3-tristetraprolin (TTP) pathways (Ronkina et al). For elucidating the probability that MAP kinase would affect ARE sequences of IL6 3’UTR to increase mRNA stability, HeLa cells infected with pCMV-UTR
15
were incubated in HBSS with MAP kinase inhibitors, SB203580, a p38 inhibitor, SP600125, a JNK inhibitor, or PD98059, an inhibitor of MEK that is a upstream signaling molecule of erk 1/2, co-treatment of SB203580 partially decreased the 3’UTR reporter actives as well as IL6 mRNA induction, co-treatment of SP600125 also showed partially decreased the reporter activities but relatively increased IL6 mRNA values. In case of PD98059 co-treatment, there is little effect on both reporter activities and IL6 induction (Fig. 9 A and B), suggesting that p38 MAP kinase-mediated regulation of 3’UTR ARE sequences influences IL6 mRNA stabilization.
16
Fig. 7. Effects of STAT3 and NF-κB transcriptional factors on IL6 3’UTR in IL6 induction of amino acid-deprived HeLa cells.
HeLa cells stably integrated with pCMV-UTR reporter plasmids, infected with non-targeting shRNA (NT), STAT3 shRNA (STAT3 sh), or p65 shRNA (p65 sh) were western-blotted for detecting expressions of STAT3, p65, or α tubulin (A). The cells were incubated in HBSS for 15 hrs, and expressions of GFP (B) and IL6 (C) mRNAs were determined by real-time PCR. Data presented as relative quantification (RQ) ± SD.
17
Fig. 8. Effects of Oncostatin M and TNF α on IL6 3’UTR in HeLa cells.
HeLa cells stably integrated with pCMV-UTR reporter plamids were incubated with HBSS or complete media containing Oncostatin M (100 ng/ml) (OSM) or TNF α (30 ng/ml) (TNF) overnight. Expression of GFP (A) and IL6 (B) mRNAs were measured by real-time PCR. Data presented as relative quantification (RQ) ± SD.
18
Fig. 9. Effects of MAP kinase inhibitor on IL6 3’UTR in amino acids-deprived HeLa cells.
HeLa cells stably infected with pCMV-UTR plasmids were incubated in HBSS in the presence of DMSO, SB203580 (10uM), SP600125 (10uM), or PD98059 (50uM). Expressions of GFP (A) and IL6 (B) mRNAs were detected by real-time PCR. Data presented as relative quantification (RQ) ± SD.
19
Ⅳ. DISCUSSION
Protein malnutrition is manifested as amino acid deprivation, which activates an amino acid response (AAR)(Kilberg et al, 2009). Amino acid limitation regulates nearly every step in gene expression, including transcription, post-transcriptional steps, and translational processes(Kilberg et al, 2005). Several of the proteins that mediate AAR-induced transcriptional activation have been identified and include members of the activating transcription factor (ATF) and CCAAT/enhancer binding protein (C/EBP) subfamilies. The general control non derepressible 2 (GCN2) kinase acts as an intracellular amino acid concentration sensor by binding uncharged tRNA, leading to phosphorylation of the translation initiation factor eukaryotic initiation factor-2α (eIF2α) on serine 51 (Sood et al, 2000). The resulting phospho-eIF2α suppresses general protein synthesis but promoters a increase in translation of selected mRNA species with upstream open reading frames, including ATF4 (Lu et al, 2004; Vattem&Wek, 2004). ATF4 triggers increased transcription from amino acid-responsive genes by binding to C/EBP-ATF response elements (CARE), so named because they are composed of a half-site for the C/EBP family and a half-site for the ATF family of transcription factors(Fawcett et al, 1999; Wolfgang et al, 1997).
GCN2 is also activation of NF-κB and STAT3 transcriptional factors, resulting in IL6 induction (Yoon et al, 2010; Yoon et al, 2011). The data showed here also has proved that NF-κB partially contribute to 3’UTR stabilization of IL6 mRNA. Supporting this, it is recently reported that NF-κB increase mRNA stability of IL6 by the pathway of Let-7 miRNA degradation which is working in 3’UTR of IL6 mRNA by way of Lin28B induction(Iliopoulos et al, 2009), and NF-κB can increase the transcription of HuR, which is a RNA-binding factor to stabilize ARE-containing mRNAs at their 3’UTR (Kang et al, 2008). Therefore, further studies elucidating association between activated NF-κB and either Let7 miRNA or HuR would be needed in future experiments during amino acid-deprivation. 3’UTR of IL6 mRNA has 6 ARE sequences extending whole 3’UTR sequences (Fig. 3).The data with deletion mutants of 3’UTR showed that serial deletion of 3’UTR decreased reporter activities but not completely, furthermore, smallest deletion mutant (pCMV-UTR d2)
20
has three ARE sequences (Fig. 5). These suggest that AREs exiting 3’UTR of IL6 mRNA are deeply implicated in mRNA stabilization after amino acid-starvation. Among RNA-binding factors, tristetraprolin (TTP) has been best studied so far, which is inactivated by phosphorylation, resulting in stabilization of TNFα mRNA via release from ARE sequences of 3’UTR. TTP can be phosphorylated by MAPK-activated protein kinases – MK2 and MK3, which is activated by p38 MAP kinase (Ronkina et al, 2010). In this context, the data showing that SB203580, an inhibitor of p38 MAP kinase partially blocks GFP-reporter activities containing IL6 3’UTR as well as IL6 mRNA after amino acid-deprivation, indicate that possibly, p38-MK2/3-TTP pathway could contribute to starvation-induced IL6 induction through stabilization of mRNA (Fig. 8). As the first step for investigating these, it would be confirmed whether p38 MAP kinase pathways are activated and how they are activated during amino acid starvation.
21
Ⅴ. CONCLUSION
1. Deprivation of amino acids induces IL6 mRNA in the presence of glucose in HeLa cells.
2. 3’UTR of IL6 is deeply implicated in this process.
3. Partially deleted 3’UTR also contributes to increase of reporter activities, suggesting that cis-acting elements within 3’UTR distribute extending whole IL6 3’UTR. 4. NF-κB knock-down partially blocks reporter activities containing 3’UTR of IL6 as
well as IL6 mRNA induction, implying that NF-κB functions in 3’UTR of IL6, subsequently stabilization of mRNA stabilization in addition to activation of IL6 promoters.
5. The studies with MAP kinase inhibitors are showing that activity of p38 MAP kinase is required for mRNA stabilization of IL6 through 3’UTR, therefore, p38-MK2/3-TTP pathway would be associated with IL6 mRNA stabilization through ARE sequences within 3’UTR sequences.
22
REFERENCES
1. Akira S, Kishimoto T (1992) The evidence for interleukin-6 as an autocrine growth factor in malignancy. Semin Cancer Biol3(1): 17-26
2. Akira S, Kishimoto T (1997) NF-IL6 and NF-kappa B in cytokine gene regulation.
AdvImmunol65: 1-46
3. Ali MH, Schlidt SA, Chandel NS, Hynes KL, Schumacker PT, Gewertz BL (1999) Endothelial permeability and IL-6 production during hypoxia: role of ROS in signal transduction. Am J Physiol277(5 Pt 1): L1057-1065
4. Anderson P, Phillips K, Stoecklin G, Kedersha N (2004) Post-transcriptional regulation of proinflammatory proteins. J LeukocBiol76(1): 42-47
5. Cheadle C, Fan J, Cho-Chung YS, Werner T, Ray J, Do L, Gorospe M, Becker KG (2005) Control of gene expression during T cell activation: alternate regulation of mRNA transcription and mRNA stability. BMC Genomics6: 75
6. Dendorfer U, Oettgen P, Libermann TA (1994) Multiple regulatory elements in the interleukin-6 gene mediate induction by prostaglandins, cyclic AMP, and lipopolysaccharide. Mol Cell Biol14(7): 4443-4454
7. Espel E (2005) The role of the AU-rich elements of mRNAs in controlling translation.
Semin Cell DevBiol16(1): 59-67
8. Fabian MR, Sonenberg N, Filipowicz W (2010) Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem79: 351-379
9. Faggioli L, Merola M, Hiscott J, Furia A, Monese R, Tovey M, Palmieri M (1997) Molecular mechanisms regulating induction of interleukin-6 gene transcription by interferon-gamma. Eur J Immunol27(11): 3022-3030
23
10. Fawcett TW, Martindale JL, Guyton KZ, Hai T, Holbrook NJ (1999) Complexes containing activating transcription factor (ATF)/cAMP-responsive-element-binding protein (CREB) interact with the CCAAT/enhancer-binding protein (C/EBP)-ATF composite site to regulate Gadd153 expression during the stress response. Biochem J339 ( Pt 1): 135-141
11. Friedman RC, Farh KK, Burge CB, Bartel DP (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res19(1): 92-105
12. Garneau NL, Wilusz J, Wilusz CJ (2007) The highways and byways of mRNA decay.
Nat Rev Mol Cell Biol8(2): 113-126
13. Iliopoulos D, Hirsch HA, Struhl K (2009) An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell139(4): 693-706
14. Isshiki H, Akira S, Tanabe O, Nakajima T, Shimamoto T, Hirano T, Kishimoto T (1990) Constitutive and interleukin-1 (IL-1)-inducible factors interact with the IL-1-responsive element in the IL-6 gene. Mol Cell Biol10(6): 2757-2764
15. Kang MJ, Ryu BK, Lee MG, Han J, Lee JH, Ha TK, Byun DS, Chae KS, Lee BH, Chun HS, Lee KY, Kim HJ, Chi SG (2008) NF-kappaB activates transcription of the RNA-binding factor HuR, via PI3K-AKT signaling, to promote gastric tumorigenesis.
Gastroenterology135(6): 2030-2042, 2042 e2031-2033
16. Kang SH, Brown DA, Kitajima I, Xu X, Heidenreich O, Gryaznov S, Nerenberg M (1996) Binding and functional effects of transcriptional factor Sp1 on the murine interleukin-6 promotor. J BiolChem271(13): 7330-7335
24
expression: how mammalian cells respond to amino acid limitation. Annu Rev Nutr25: 59-85
18. Kilberg MS, Shan J, Su N (2009) ATF4-dependent transcription mediates signaling of amino acid limitation. Trends EndocrinolMetab20(9): 436-443
19. Libermann TA, Baltimore D (1990) Activation of interleukin-6 gene expression through the NF-kappa B transcription factor.Mol Cell Biol10(5): 2327-2334
20. Lu PD, Harding HP, Ron D (2004) Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J Cell Biol167(1): 27-33 21. Lust JA (1994) Role of cytokines in the pathogenesis of monoclonal
gammopathies.Mayo ClinProc69(7): 691-697
22. Ronkina N, Menon MB, Schwermann J, Tiedje C, Hitti E, Kotlyarov A, Gaestel M (2010) MAPKAP kinases MK2 and MK3 in inflammation: complex regulation of TNF biosynthesis via expression and phosphorylation of tristetraprolin.
BiochemPharmacol80(12): 1915-1920
23. Sehgal PB (1990) Interleukin 6 in infection and cancer. ProcSocExpBiol Med195(2): 183-191
24. Shi Y, Porter K, Parameswaran N, Bae HK, Pestka JJ (2009) Role of GRP78/BiP degradation and ER stress in deoxynivalenol-induced interleukin-6 upregulation in the macrophage. ToxicolSci109(2): 247-255
25. Sood R, Porter AC, Olsen DA, Cavener DR, Wek RC (2000) A mammalian homologue of GCN2 protein kinase important for translational control by phosphorylation of eukaryotic initiation factor-2alpha. Genetics154(2): 787-801
25
26. Tanaka M, Miyajima A (2003) Oncostatin M, a multifunctional cytokine. Rev
PhysiolBiochemPharmacol149: 39-52
27. Vattem KM, Wek RC (2004) Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. ProcNatlAcadSci U S A101(31): 11269-11274 28. vonRoretz C, Gallouzi IE (2008) Decoding ARE-mediated decay: is microRNA part of
the equation? J Cell Biol181(2): 189-194
29. Wolfgang CD, Chen BP, Martindale JL, Holbrook NJ, Hai T (1997) gadd153/Chop10, a potential target gene of the transcriptional repressor ATF3. Mol Cell Biol17(11): 6700-6707
30. Xiao G, Fu J (2011) NF-kB and cancer: a paradigm of Yin-Yang. Am J Cancer Res1(2): 192-221
31. Yamauchi-Takihara K, Ihara Y, Ogata A, Yoshizaki K, Azuma J, Kishimoto T (1995) Hypoxic stress induces cardiac myocyte-derived interleukin-6. Circulation91(5): 1520-1524
32. Yoon S, Woo SU, Kang JH, Kim K, Kwon MH, Park S, Shin HJ, Gwak HS, Chwae YJ (2010) STAT3 transcriptional factor activated by reactive oxygen species induces IL6 in starvation-induced autophagy of cancer cells. Autophagy6(8): 1125-1138
33. Yoon S, Woo SU, Kang JH, Kim K, Shin HJ, Gwak HS, Park S, Chwae YJ (2011) NF-kB and STAT3 cooperatively induce IL6 in starved cancer cells. OncogeneIn press
26 -국문요약-
HeLa
세포주의 아미노산 결핍에서 IL6 유전자의 3’ UTR이
mRNA의 표현에 미치는 영향 분석
아주대학교 대학원 의생명과학과
강 정 희
(지도 교수 : 최 용 준)
암세포의 아미노산 결핍은 STAT3와 NF-kB 전사 인자를 활성화 시킴으로 인해 ER stress responses 와 autophagic responses를 발생시키고 IL6 사이토카인을 유도 한 다. 세포 자극반응을 통하여 IL6 사이토카인이 유도됨을 IL6 mRNA의 3’UTR의 역할을 통해 연구 하였다. IL6 mRNA의 GFP-reporter를 포함한 3’UTR의 연구에 서, 아미노산 결핍 후 IL6 mRNA의 증가에 3’UTR이 관여하고, 이 현상들은 IL6 3’UTR의 결실돌연변이(Deletion mutation)의 형태에 따라서도 광범위하게 나타내어 진다.
NF-κB subunit인 p65의 knock-down 실험에서 3’UTR은 부분적으로 IL-6 mRNA의 안정성에 관여하는 것으로 보여진다. 그리고 p38 MAP kinase inhibitor SB203580을 처리 하였을 때 아미노산 결핍에 따른 IL6 감소뿐 아니라 3’UTR을 통한 mRNA 의 안정성을 현저하게 저해했다. 이것은 p38-MK2/3-TTP pathway가 암세포주의 영
27
양분 고갈 상태에서 IL6 mRNA의 안정성과 관련이 있고, 활성화 영향을 준다는 것을 제시한다.
결론적으로 이 연구는 NF-κB와 p38에 관한 stress signal은 3’UTR sequence를 통 해 mRNA 안정성에 관여하지만, 이것에 대한 분자적 구조에 대한 연구는 더 필 요 할 것이다.
핵심어 : IL6 cytokines, AU rich element, 3’untranslation region, post-transcription, mRNA stability and translation