저작자표시-비영리-동일조건변경허락 2.0 대한민국 이용자는 아래의 조건을 따르는 경우에 한하여 자유롭게 l 이 저작물을 복제, 배포, 전송, 전시, 공연 및 방송할 수 있습니다. l 이차적 저작물을 작성할 수 있습니다. 다음과 같은 조건을 따라야 합니다: l 귀하는, 이 저작물의 재이용이나 배포의 경우, 이 저작물에 적용된 이용허락조건 을 명확하게 나타내어야 합니다. l 저작권자로부터 별도의 허가를 받으면 이러한 조건들은 적용되지 않습니다. 저작권법에 따른 이용자의 권리는 위의 내용에 의하여 영향을 받지 않습니다. 이것은 이용허락규약(Legal Code)을 이해하기 쉽게 요약한 것입니다. Disclaimer 저작자표시. 귀하는 원저작자를 표시하여야 합니다. 비영리. 귀하는 이 저작물을 영리 목적으로 이용할 수 없습니다. 동일조건변경허락. 귀하가 이 저작물을 개작, 변형 또는 가공했을 경우 에는, 이 저작물과 동일한 이용허락조건하에서만 배포할 수 있습니다.
Production of
recombinant monoclonal antibodies
against HSET
by
Youngsil Seo
Major in Molecular Medicine
Department of Biomedical Sciences
The Graduate School, Ajou University
Production of
recombinant monoclonal antibodies
against HSET
by
Youngsil Seo
A Dissertation Submitted to The Graduate School of
Ajou University in Partial Fulfillment of the Requirements
for the Degree of Master of Biomedical Sciences
Supervised by
Myung-Hee Kwon, Ph.D.
Major in Molecular Medicine
Department of Biomedical Sciences
The Graduate School, Ajou University
i - ABSTRACT -
Production of recombinant monoclonal antibodies against HSET
HSET (Human Spleen Embryo Testis), belonged to the kinesin 14A family, is a protein required
for centrosome clustering during cell division. Since cancer cells with extra-centrosomes could
lead to multi-polar division, resulting in genomic instability and cell death, these cancer cells
depend on the centrosome clustering activity of HSET that helps normal chromosome
segregation by focusing several centrosomes into two poles. As the first step of the strategy to
inhibit the function of HSET in cancer cells, we produced mouse monoclonal antibodies (mAbs)
against HSET using a hybridoma technique and characterized them. Peptide antigens of 12
amino acid (aa)-length, designed from the regions corresponding to N-terminus (1-134th aa) and
C-terminus (626-672th aa) that do not have homology with other kinesin molecules, were used
for mouse immunization. Total 28 of mAbs against the peptides from N-terminus HSET were
obtained, but no mAb against C-terminus of HSET. In ELISA with the Abs in ascitic fluids
against yeast surface displayed HSET, clone 8C346 showed high affinity. In ELISA using Abs
purified from hybridoma culture supernatants, clone 1C274, 2C280, 2C281, 6C407, 9C352, and
9C353 showed relatively strong binding to Whole HSET protein. We expect that Abs with high
binding to HSET could inhibit HSET function, resulting in selective death of cancer cells.
ii
TABLE OF CONTENTS
ABSTRACT ··· i TABLE OF CONTENTS ··· ii LIST OF FIGURES ··· iv LIST OF TABLES ··· vi Ⅰ. INTRODUCTION ··· 1Ⅱ. MATERIALS AND METHODS ··· 4
A. Production of anti-HSET mAbs secreting hybridoma cell line ··· 4
B. Confirmation of mAbs in ascitic fluids by western blotting ··· 4
C. Displaying HSET fragment on the surface of yeast cells ··· 5
D. Preparation of whole HSET protein ··· 5
E. Enzyme-linked Immunosorbent Assay (ELISA) for binding of mAbs to HSET ·· 6
F. Sequence analysis of variable region gene in immunoglobulin from hybridoma cell lines ··· 7
iii
Ⅲ. RESULTS ··· 9
A. The design of peptide antigen for immunization ··· 9
B. Preparation of antigen ··· 15
C. Binding of anti-HSET mAbs to different forms of HSET antigen ··· 18
D. Comparison of binding activity between mAbs ··· 26
E. Sequence analysis of anti-HSET mAbs ··· 29
IV. DISCUSSION ··· 32
V. CONCLUSION ··· 34
REFERENCES ··· 35
iv
LIST OF FIGURES
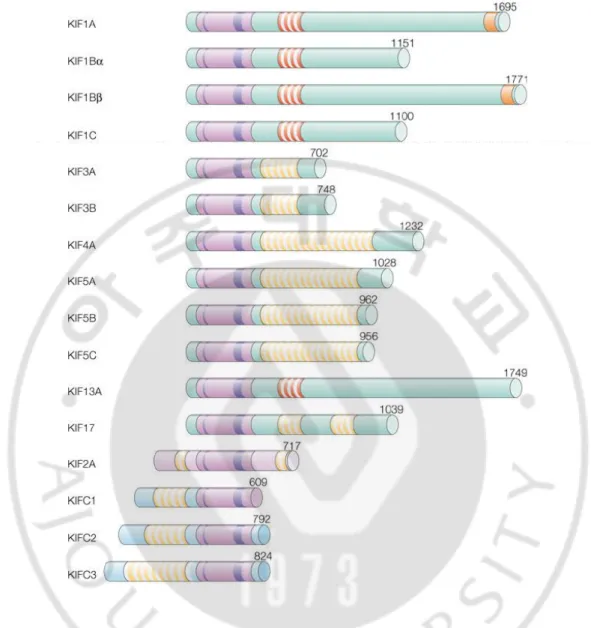
Figure 1. Structures of kinesins ··· 10
Figure 2. Amino acids sequence of human HSET ··· 11
Figure 3. BLAST search of whole HSET protein (Met1 - Lys673) sequence··· 12
Figure 4. Confirmation of yeast surface displayed HSET fragment ··· 16
Figure 5. Purification of whole HSET (Met1 - Lys673) protein ··· 17
Figure 6. Western blotting of the ascitic fluids containing anti-HSET mAbs ··· 20
Figure 7. ELISA for the binding of ascitic fluids containing anti-HSET mAbs to peptide used for immunization ··· 21
Figure 8. ELISA for the binding of ascitic fluids containing anti-HSET mAbs to synthetic HSET peptide (Met1 - Lys50) ··· 22
Figure 9. ELISA for the binding of ascitic fluids containing anti-HSET mAbs to yeast displayed HSET fragment (Met1 - Ala100) ··· 23
v
Figure 11. ELISA for the binding of the purified anti-HSET mAbs
to whole HSET protein antigen ··· 25
vi
LIST OF TABLES
Table 1. Primers used for amplifying variable region genes of hybridoma cell lines ·· 8
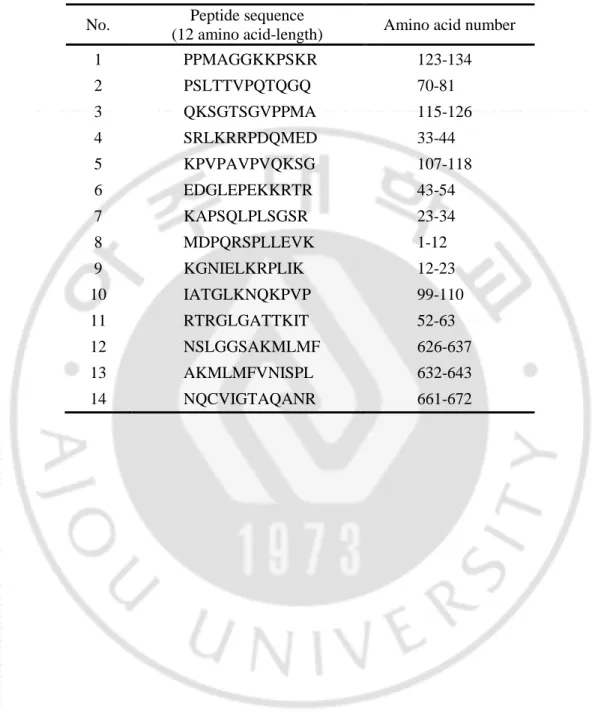
Table 2. Peptides sequence of HSET used for immunization ··· 13
Table 3. The location of peptide antigens used for immunization ··· 14
Table 4. Summary of anti-HSET mAbs ··· 27
1
I. INTRODUCTION
The centrosome is an important cell organelle that participates in spindle assembly and
chromosome segregation during mitosis (Doxsey, 2001). Duplication of centrosome is tightly
regulated once per cell cycle, thus cells in mitosis should have two functional centrosomes
(Sharp et al., 2000). Unlike normal cells, some cancer cells have amplified number of
centrosomes. It is still controversial whether amplified centrosomes are the consequence or
beginning of tumorigenesis (Boveri, 2008), but cancer cells have been evolved to achieve
bi-polar division with extra-centrosomes. Several mechanisms are known to avoid multi-bi-polar
division such as extrusion, inactivation, segregation, and clustering of extra-centrosomes
(Manandhar et al., 2005; Basto et al., 2008; Godinho et al., 2009).
Since it was observed in N115 mouse neuroblastoma cell line (Ring et al., 1982) and breast
tissue (Salisbury et al., 1999), centrosome clustering mechanism has been studied. Microtubules
binding proteins such as integrin-linked kinase (ILK), dynein, and HSET are related to
clustering of centrosomes in different mechanisms (Ogden et al., 2012). ILK localized at
centrosome regulates the formation of mitotic spindle (Fielding et al., 2008). Dynein, minus end
directed motor protein, recruits nuclear mitotic apparatus protein (NuMa) to spindle poles and
bundles centrosomes (Quintyne et al., 2005). Over-expression of NuMa interrupts the function
of dynein, resulting in declustering of centrosomes.
HSET belongs to the kinesin 14A family and consists of microtubule binding domain,
coiled coil domain for homo-dimer formation, and motor domain. It has been known that HSET
2
triphosphate (ATP) by motor domain. Binding and sliding between microtubule, HSET moves
to the spindle pole and gathers extra-centrosomes into two poles. During interphase, HSET is
sequestered in the nucleus (Cai et al., 2009). As cells enter mitosis, HSET spreads out cytosol
and binds to microtubule after nuclear membrane breakdown. The HSET level of mRNA was
significantly increased in tumor tissues than normal tissues (Kleylein-Sohn et al., 2012). In the
absence of HSET, the viability of cancer cells that mostly contain extra-centrosomes was
decreased about 50%. In contrast, cancer cell lines containing two centrosomes showed only
slightly reduced cell viability (Kwon et al., 2008). Centrosome clustering activity of HSET
makes it an attractive anti-cancer target molecule because the role of HSET is dispensable in
normal cells (Kleylein-Sohn et al., 2012).
Focused on the difference between normal and cancer cells in dependency to HSET, there
were several trials to inhibit centrosome clustering in cancer cells using small molecule
inhibitors. For example, griseofulvin, which has been used as an antifungal agent clinically,
induces multi-polar division in cancer cells (Rebacz et al., 2007; Raab et al., 2012). It is known
that griseofulvin binds to microtubule and inhibits its function. Noscapaine and its derivative
(brominated noscapine) are known to de-cluster centrosomes, accompanying G2/M arrest and
apoptotic cell death (Aneja et al., 2006; Karna et al., 2011). Phenanthrene-derived poly
ADP-ribose polymerase (PARP) inhibitor is also known as a centrosome de-clustering agent (Castiel
et al., 2011).
Until now, two synthetic compounds have been reported to inhibit the activity of HSET in
cancer cells (Watts et al., 2013; Wu et al., 2013). One is CW069 that can selectively recognize
HSET and bind to the loop 5 cleft in the HSET motor domain. CW069 can change the structure
of ATP binding site in HSET by binding in an allosteric manner. In extra-centrosome containing
3
treatment of CW069, similar extent induced by knock-down of HSET using siRNA. The other
compound, AZ82, can inhibit the enzymatic activity of HSET in an ATP-competitive manner.
Nevertheless several advantages of centrosome de-clustering drugs, it is preferred to use
monoclonal antibody (mAb) as a therapeutic agent because of safety and half-life in vivo
(Chames et al., 2009; Goldenberg and Sharkey, 2012). Injection of anti-HSET mAbs into cells
induced the abnormal movement of chromosome (Gordon et al., 2001), suggesting that HSET
function can be inhibited by Ab.
The purpose of this study is to generate anti-HSET mAbs that could inhibit HSET function
in cancer cells. Mouse anti-HSET mAbs were produced using a hybridoma technique and then
4
II. MATERIALS AND METHODS
A. Production of anti-HSET mAbs secreting hybridoma cell line
Peptide antigen were immunized to three 8 to 12-week old BALB/C mice (female) and the
mice that showed high immune response were chosen. Mouse spleen cells were fused with
SP2/0 myeloma cells to generate hybridoma. Cells were then diluted and clones were grown
from single parent cells on microtiter wells in hypoxanthine-aminopterin-thymidine (HAT)
medium. The mAbs secreted by the different clones were assayed for their ability to bind to the
antigen by ELISA. The most productive and stable clones were selected and injected in the
peritoneal cavity of mice. After 10-14 days, antibody-rich fluids called ascitic fluids were
collected from mice. High purity mAbs were also purified from culture supernatant of
hybridoma cells using protein G resin for assays.
B. Confirmation of mAbs in ascitic fluids by western blotting
Ascitic fluids were separated on 12% acrylamide gels with 3 µl per well and transferred
onto polyvinylidene difluoride (PVDF) membrane. After blocking of the membrane with 5%
skim milk in TBST (0.1% Tween20 in 10 mM Tris-Cl, 150 mM NaCl, pH 7.2) for 2 hrs at room
temperature, the membrane was incubated with 10,000-fold diluted alkaline phosphatase
(AP)-conjugated goat anti-mouse IgG (Fc specific) mAbs (Sigma) at 4°C overnight to detect heavy
5
developed by 5-Bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium (BCIP/NBT) for 5
min.
C. Displaying HSET fragment on the surface of yeast cells
HSET fragment (Met1 - Ala100) was amplified using restriction enzyme site-containing
primers (HsNtNheI 5’GATCTAGCTAGCATGGACCCGCAGAGGTCCCCCCTATTGG
-3’, Hs-Nt-BamHI 5’- GAGATCAGTCTCGAGCTTCCTGTTGGCCTGAGC -3’). PCR
products were digested with BamHI and NheI, then inserted into yeast surface display vector
pCTCON and transformed into yeast strain EBY100 by electroporation. Yeast cells were
grown in SD-CAA at 30°C until absorbance at 600nmreaches 6.0 and the media were changed
into SG-CAA to induce expression of HSET fragment at 20°C for 20 hrs. To confirm the
expression level of HSET fragment on yeast cell surface, yeast cells were washed with TBST
and labeled with mouse anti-myc mAb (IG THERAPY, IG-A03002) for 30 min at room
temperature. After washing cells with TBST once, yeast cells were labeled with
FITC-conjugated goat anti-mouse IgG as a secondary Ab (Sigma, A1418) for 15 min at 4°C. Then,
yeast cells were washed with TBST once, followed by flow cytometric analysis.
D. Preparation of whole HSET protein
HSET (Met1 - Lys673) gene (ORIGENE, RG215326) was amplified by PCR using
restriction enzyme site-containing primers (BamHI-HSET 5’-
GATCTAGGATCCATGGACCCGCAGAGG -3’, HSET-XhoI 5’-
GAGATCAGTCTCGAGCTTCCTGTTGGCCTGAGC -3’) and Pfu polymerase (Intron, Korea).
6
expression vector (Novagen) to produce whole HSET protein. Escherichia coli BL21 (DE3)
pLysE cells (Novagen) were transformed with pET21a-HSET plasmid and grown in 1 L of
Luria Bertani medium with 100 μg/ml of ampicillin and 25 μg/ml of chloramphenicol at 30°C,
until absorbance reaches 0.6 at 600 nm. Expression of whole HSET protein was induced by
adding 0.5 mM of isopropyl 1-thio-β-D-galactopyranoside.
After growing cells at 25°C for 6 hrs, cells were collected by centrifugation at 8000 rpm for
30 min at 4°C. By grinding liquid nitrogen-frozen pellet using mortar and pestle, cell walls
could be disrupted. Pellet powder was re-suspended with 30 ml of binding buffer (0.5% NP-40
in 50 mM Tris-HCl, 300 mM NaCl, pH 8.0) containing EDTA-free protease inhibitor cocktail
(Roche). After mixing the mixture in rotator for 15 min at 4°C, cells were sonicated in 30% of
amplitude, 20 sec pulse on, 30 sec pulse off, for total 1 min 20 sec processing time.
Supernatant was collected by centrifugation at 8000 rpm for 30 min at 4°C. 2 ml of cobalt
resin was mixed with the supernatant at 4°C for 2 hrs. Cobalt resin was transferred to column
and washed with washing buffer (20 mM of imidazole in binding buffer) several times. Whole
HSET protein was eluted by adding 20 ml of elution buffer (300 mM imidazole in binding
buffer) to column and collected in Vivaspin (molecular weight cut-off 10,000 Dalton). Whole
HSET protein was concentrated by centrifugation at 5000 rpm at 4°C and then buffer was
changed to TBS.
E. Enzyme-linked Immunosorbent Assay (ELISA) for binding of mAbs to HSET
Ninety six-well polystyrene microtiter plate (Nunc, Invitrogen) were coated with
immunogen (0.05 μg/well) Met1 - Lys50 of HSET synthetic peptide (0.1 μg/well) or HSET
7
Lys673) protein (2.5 μg/well) at 4°C overnight. The wells were blocked with 5% skimmed milk
for 2 hr at room temperature. After discard blocking solution by tapping, anti-HSET mAbs
(50-fold diluted ascitic fluids, 5 µg of purified mAbs in blocking solution) were treated for 1 hr at
room temperature. The wells were washed with TBST three times and treated with
AP-conjugated goat anti-mouse IgG (Sigma) for 1 hr at room temperature. The wells were washed
with TBST three times and ρ-nitrophenyl phosphate solution (1 mg/ml in 0.1 M glycine, 1 mM
ZnCl2, 0.1 mM MgCl2, pH 10.4) was added to the wells as a substrate of secondary Ab. The
absorbance was read at 405 nm in a microplate reader.
F. Sequence analysis of variable region gene in immunoglobulin from hybridoma cell lines
Total RNA from each hybridoma cell lines was extracted using easy-BLUE Total RNA
Extraction Kit (Intron, Korea) and cDNA was synthesized by PrimeScript RT Master Mix
(Takara) using total RNA as a template. Variable region genes were amplified by polymerase
chain reaction (PCR) using primers that specifically bind to framework or constant regions of
immunoglobulin genes described in Table 1 (Williams et al., 2010). Sequences of PCR products
8
Table 1. Primers used for amplifying variable region genes of hybridoma cell lines
Name Sequencee MHV1a ATGAAATGCAGCTGGGGCATSTTCTTC MHV2 ATGGGATGGAGCTRTATCATSYTCTT MHV3 ATGAAGWTGTGGTTAAACTGGGTTTTT MHV4 ATGRACTTTGGGYTCAGCTTGRTTT MHV5 ATGGACTCCAGGCTCAATTTAGTTTTCCTT MHV6 ATGGCTGTCYTRGSGCTRCTCTTCTGC MHV7 ATGGRATGGAGCKGGRTCTTTMTCTT MHV8 ATGAGAGTGCTGATTCTTTTGTG MHV9 ATGGMTTGGGTGTGGAMCTTGCTATTCCTG MHV10 ATGGGCAGACTTACATTCTCATTCCTG MHV11 ATGGATTTTGGGCTGATTTTTTTTATTG MHV12 ATGATGGTGTTAAGTCTTCTGTACCTG Cɤ Ib GGACAGGGATCCAGAGTTCCA Cɤ II TGGATAGACAGATGGGGGTGTCGTTTTGGC MKV1c AAGTTGCCTGTTAGGCTGTTGTGTCTC MKV2 ATGGAGWCAGACACACTCCTGYTATGGGTG MKV3 ATGAGTGTGCTCACTCAGGTCCTGGSGTTG MKV4 ATGAGGRCCCCTGCTCAGWTTYTTGGMWTCTTG MKV5 ATGGATTTWCAGGTGCAGATTWTCAGCTTC MKV6 ATGAGGTKCYYTGYTSAGYTYCTGRGG MKV7 ATGGGCWTCAAGATGGAGTCACAKWYYCWGG MKV8 ATGTGGGGAYCTKTTTYCMMTTTTTCAATTG MKV9 ATGGTRTCCWCASCTCAGTTCCTTG MKV10 ATGTATATATGTTTGTTGTCTATTTCT MKV11 ATGGAAGCCCCAGCTCAGCTTCTCTTCC Cĸ Id TGAGGCACCTCCAGATGTTAA Cĸ II GGATGGTGGGAAGATGGATAC Cĸ III AGTTGGTGCAGCATCAGC a
MHV and bCɤ indicate primers that anneal to leader sequences of mouse heavy chain variable and constant region genes.
c
MKV and dCĸ indicate primers that anneal to leader sequences of mouse kappa light chain variable and constant region genes.
e
Ambiguity code: M = A or C; R = A or G; W = A or T; S = C or G; Y = C or T; K = G or T
9
III. RESULTS
A. The design of peptide antigen for immunization
HSET belongs to kinesin family (Hirokawa and Takemura, 2005) and most of kinesin
proteins share similar sequences of amino acid with HSET (Fig. 1). Amino acid sequence and
each domain of HSET are shown in Figure 2. To increase specificity of Ab to HSET, it is
essential to avoid targeting the regions which show high homology with other kinesins. We
performed BLAST search for HSET to avoid cross-reactivity of anti-HSET Ab with kinesin
proteins (Fig. 3). N-terminus (Met1 - Arg134) and C-terminus (Asn626 - Arg672) regions of
HSET showed relatively low homology to other kinesins, compared to motor region of HSET.
The coiled coil region (Glu249 - Glu306) was considered to be unsuitable to use as antigen
because it has many hydrophobic amino acids and moderate similarity with other kinesins.
Finally we designed 12 amino acid-length peptides from 14 hydrophilic regions
corresponding to N- and C-terminus of HSET for mice immunization (Table 2) and the location
10
Figure 1. Structures of kinesins. The structures of the kinesin family. The motor domains (pink-colored), the ATP-binding site (a thin purple line), the microtubule-binding site (a thick
purple line), the dimerization domains (yellow stripes), the forkhead-associated regions (red
stripes) and pleckstrin homology domains (orange stripes) are shown as schematic diagram. The
number of amino acids of each molecule is shown on the right. A partial region (1st - 609th
11
10 20 30 40 50 60 MDPQRSPLLE VKGNIELKRP LIKAPSQLPL SGSRLKRRPD QMEDGLEPEK KRTRGLGATT
70 80 90 100 110 120 KITTSHPRVP SLTTVPQTQG QTTAQKVSKK TGPRCSTAIA TGLKNQKPVP AVPVQKSGTS
130 140 150 160 170 180 GVPPMAGGKK PSKRPAWDLK GQLCDLNAEL KRCRERTQTL DQENQQLQDQ LRDAQQQVKA
190 200 210 220 230 240
LGTERTTLEG HLAKVQAQAE QGQQELKNLR ACVLELEERL STQEGLVQEL QKKQVELQEE
250 260 270 280 290 300
RRGLMSQLEE KERRLQTSEA ALSSSQAEVA SLRQETVAQA ALLTEREERL HGLEMERRRL
310 320 330 340 350 360
HNQLQELKGN IRVFCRVRPV LPGEPTPPPG LLLFPSGPGG PSDPPTRLSL SRSDERRGTL 370 380 390 400 410 420
SGAPAPPTRH DFSFDRVFPP GSGQDEVFEE IAMLVQSALD GYPVCIFAYG QTGSGKTFTM
430 440 450 460 470 480
EGGPGGDPQL EGLIPRALRH LFSVAQELSG QGWTYSFVAS YVEIYNETVR DLLATGTRKG
490 500 510 520 530 540
QGGECEIRRA GPGSEELTVT NARYVPVSCE KEVDALLHLA RQNRAVARTA QNERSSRSHS
550 560 570 580 590 600
VFQLQISGEH SSRGLQCGAP LSLVDLAGSE RLDPGLALGP GERERLRETQ AINSSLSTLG
610 620 630 640 650 660
LVIMALSNKE SHVPYRNSKL TYLLQNSLGG SAKMLMFVNI SPLEENVSES LNSLRFASKV
670 NQCVIGTAQA NRK
Figure 2. Amino acids sequence of human HSET. Amino acid from 36 to 52th (orange-colored) is a nuclear localization signal which makes HSET localizing at nuclear. Coiled coil
region (142-306th amino acid, pink-colored) are related to dimerization of HSET. Motor
domain corresponding to 307-594th amino acid (blue-colored) has nucleotide binding site
(410-417th amino acid, cyan-colored) and ATP hydrolyzing activity. Underlined italic letters indicate
12
Figure 3. BLAST search of whole HSET protein (Met1 - Lys673) sequence. Whole HSET protein sequence was analyzed using BLAST to know which region has similarity with other
kinesin proteins. The red bar on the top indicates whole HSET protein against in which
13
Table 2. Peptides sequence of HSET used for immunization
No. Peptide sequence
(12 amino acid-length) Amino acid number
1 PPMAGGKKPSKR 123-134 2 PSLTTVPQTQGQ 70-81 3 QKSGTSGVPPMA 115-126 4 SRLKRRPDQMED 33-44 5 KPVPAVPVQKSG 107-118 6 EDGLEPEKKRTR 43-54 7 KAPSQLPLSGSR 23-34 8 MDPQRSPLLEVK 1-12 9 KGNIELKRPLIK 12-23 10 IATGLKNQKPVP 99-110 11 RTRGLGATTKIT 52-63 12 NSLGGSAKMLMF 626-637 13 AKMLMFVNISPL 632-643 14 NQCVIGTAQANR 661-672
14
15 B. Preparation of antigen
To confirm the binding activity of mAbs with expanded length antigens instead of 12
amino acid-length short peptide, we prepared antigen by various methods. Firstly, we
synthesized a 50 amino acid-length peptide corresponding to N-terminus of HSET (Met1 -
Lys50).
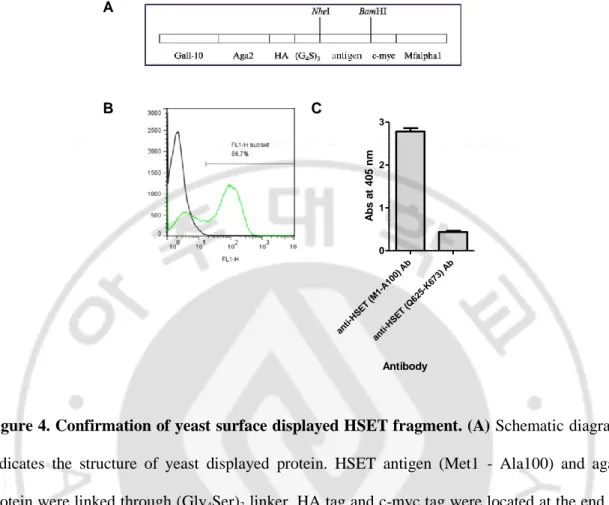
Also a little bit longer, HSET (Met1 - Ala100) proteins were fused with yeast surface
protein Aga2 using yeast expression vector and displayed on the surface of yeast cells (Fig. 4A).
The expression level of HSET fragment displayed on the yeast was detected using c-myc tag
located at C-terminus of HSET and FITC conjugated secondary Ab through flow cytometry (Fig.
4B). As a result, more than 60% of yeast cells successfully displayed HSET fragment. To use
yeast surface displayed protein as an antigen of ELISA, we determined whether yeast cells
could be coated on the 96-well microtiter plate or not. As a result, it is confirmed that yeast cells
could be coated on the plate and detected by anti-HSET Ab (Fig. 4C).
To ascertain that mAbs bind to full-length protein, whole HSET protein tagged with
hexa-histidine at the C-terminus was purified from E.coli. Protein in cell lysates was captured by
cobalt resin via histidine tag and eluted by addition of high concentration imidazole solution.
We obtained 700 µg of protein per 1 L culture. The purity of protein was confirmed by
SDS-PAGE and western blotting (Fig. 5). The bands below 50 kDa detected by western blotting were
thought to be the degraded forms of HSET. The band near 70 kDa (Fig. 5A) seems to be HSET
protein in which N-terminus and C-terminus are lost, because it was not detected by both
16
A
B C
Figure 4. Confirmation of yeast surface displayed HSET fragment. (A) Schematic diagram indicates the structure of yeast displayed protein. HSET antigen (Met1 - Ala100) and aga2
protein were linked through (Gly4Ser)3 linker. HA tag and c-myc tag were located at the end of
aga2 and antigen, respectively. (B) Yeast displayed Aga2-HSET fusion protein are monitored
by flow cytometry. Using anti-myc mAbs and FITC conjugated secondary Ab, it is found that
more than 60% of yeast cells were expressing HSET fragment. (C) HSET fragment displaying
yeast cells were coated on the 96-well plate and detected by HSET specific mAbs through
ELISA. ant i-HS ET (M1 -A1 00) Ab ant i-HS ET (Q 62 5-K673 ) A b 0 1 2 3 Antibody A b s a t 4 0 5 n m
17
A B C
Figure 5. Purification of whole HSET (Met1 - Lys673) protein. (A) SDS-PAGE of the whole HSET protein (74 kDa) purified by affinity chromatography using a cobalt resin. Protein (10
μg) were loaded on 10% acrylamide gels under denature condition and visualized with Coomassie Blue. (B, C) Western blottings. The purified whole HSET protein was detected with
either mouse anti-His tag mAb (B) or mouse anti-HSET (the region of Met1 - Lys50) mAb (C),
18
C. Binding of anti-HSET mAbs to different forms of HSET antigen
Twenty-eight hybridoma cell lines were generated using a hybridoma technique with
immunization of HSET peptides (Table 2). To get high concentration of mAb, we injected
hybridoma cells into abdominal cavity of mice and collected the Ab-rich ascitic fluids. On the
other hands, Abs were purified from the supernatant of hybridoma cells by affinity
chromatography. The ascitic fluids and the Abs purified from hybridoma culture supernatant
were analyzed by western blotting and SDS-PAGE (Fig. 6 and 10).
Four different forms of HSET antigens were prepared to analyze the binding activity of
twenty-eight anti-HSET mAbs. Three antigens including 12 amino acid-length peptides used for
mice immunization, 50 amino acid-length synthetic peptide (Met1 - Lys50), and 100 amino
acid-length HSET fragment (Met1 - Ala100) displayed on the yeast surface were used for
ELISA to analyze the binding activities of mAbs in ascitic fluids. The purified whole HSET
antigen was used to analyze the binding of the purified IgG mAbs.
First, we analyzed the binding of mAbs in ascitic fluids to 12 amino acid-length peptides.
Most of clones bound to each peptide which had been used for immunization with high extent,
except for clone 1C272, 2C279, 2C280, and 2C281 (Fig. 7). To confirm whether mAbs in
ascitic fluids bind to longer synthetic peptide (Met1 - Lys50), ELISA was performed.
Anti-HSET mAbs in ascitic fluids were incubated with the peptide-coated wells and then detected
with AP-conjugated anti-mouse IgG Ab. Clone 6C407, 8C346, 9C352, and 9C353 showed high
binding, compare to positive control that commercial anti-HSET mAb (Fig. 8). MAbs in ascitic
fluids, which target epitopes between Met1 - Ala100 of HSET, were analyzed by ELISA using
19
high binding and clone 6C407 and 7C315 bound 1.7-fold greater than negative control to HSET
N-terminus (Met1 - Ala100) displayed on yeast cells (Fig. 9).
In ELISA using ascitic fluids, 2 µl of ascitic fluids was treated per well. Considering that
the amount of mAbs in 3 µl of ascitic fluids was similar to 1 µg of positive control mouse IgG
(Fig. 6), about 1 µg of mAbs were treated to each well. Although concentration of mAbs was
different between clones, it seems that binding of mAbs to antigen was mainly determined by
the affinity of mAbs. It was because that 8C346 Ab amount in ascitic fluids was smaller than
8C344, but 8C346 showed higher binding to 50 amino acid-length peptide and yeast displayed
HSET fragment in ELISA (Fig. 8 and 9).
When the purified mAbs were stored in 50% of glycerol/ 0.05% of sodium azide,
aggregation of the purified mAbs was observed, making us unable to determine accurate
concentration of the purified mAbs. The concentration of the purified mAbs used for ELISA
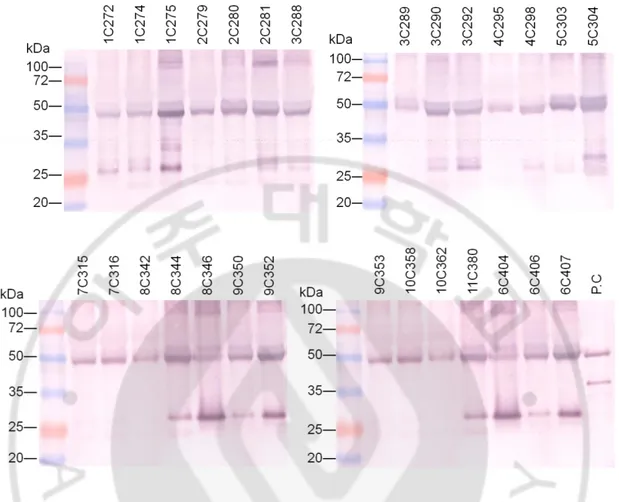
was compensated based on SDS-PAGE result (Fig. 10). The binding activity of the purified
mAbs to whole HSET (Met1 - Lys673) protein was confirmed by ELISA. Clone 1C274, 2C280,
2C281, 6C407, 9C352, and 9C353 bound to whole HSET protein, to similar extent with a
20
Figure 6. Western blotting of the ascitic fluids containing anti-HSET mAbs. The aliquots (3 µl) of ascitic fluids that had been obtained by i.p. injection of the hybrioma cells were run on
12% acrylamide gels under denature condition. Then, the mAbs were detected by Western
blotting using AP-conjugated goat anti-mouse IgG (Fc-specific). Pure mouse IgG of 1µg was
21 1 C2 7 2 1 C2 7 4 1 C2 7 5 2 C2 7 9 2 C2 8 0 2 C2 8 1 3 C2 8 8 3 C2 8 9 3 C2 9 0 3 C2 9 2 4 C2 9 5 4 C2 9 8 5 C3 0 3 5 C3 0 4 6 C4 0 4 6 C4 0 6 6 C4 0 7 7 C3 1 5 7 C3 1 6 8 C3 4 2 8 C3 4 4 8 C3 4 6 9 C3 5 0 9 C3 5 2 9 C3 5 3 1 0 C3 5 8 1 0 C3 6 2 1 1 C3 0 8 N.C PBS 0.0 0.5 1.0 1.5 2.0 2.5
Immunogen peptide coating PBS coating Hybridoma clone A b s a t 4 0 5 n m
Figure 7. ELISA for the binding of ascitic fluids containing anti-HSET mAbs to peptide used for immunization. Eleven kinds of immunogen peptides (0.05 μg/well) were coated on the 96-well plate, followed by treatment of 50-fold diluted ascitic fluids. MAbs that bound to
immunogen were detected by AP conjugated goat anti-mouse IgG. As negative controls,
irrelevant ascitic fluid (N.C) or PBS was used. The first digit of the clone name stands for
22 4 C 2 9 5 4 C 2 9 8 6 C 4 0 4 6 C 4 0 6 6 C 4 0 7 7 C 3 1 5 7 C 3 1 6 8 C 3 4 2 8 C 3 4 4 8 C 3 4 6 9 C 3 5 0 9 C 3 5 2 9 C 3 5 3 P .C N .C PBS 0.0 0.5 1.0 1.5 2.0 2.5 Hybridoma clone A b s a t 4 0 5 n m
Figure 8. ELISA for the binding of ascitic fluids containing anti-HSET mAbs to synthetic HSET peptide (Met1 - Lys50). Synthetic HSET peptide (1 µg/ml) was coated on 96-well plate and then 50-fold diluted ascitic fluids were treated. MAbs were detected by AP conjugated goat
anti-mouse IgG. As negative controls, irrelevant ascitic fluid (N.C) or PBS was used.
Commercial anti-HSET mAb (from Abcam) was used as a positive control. The first digit of the
clone name stands for peptide antigen number. Data represent the mean ± S.D. of triplicate
23 2 C 2 7 9 2 C 2 8 0 2 C 2 8 1 4 C 2 9 5 4 C 2 9 8 6 C 4 0 4 6 C 4 0 6 6 C 4 0 7 7 C 3 1 5 7 C 3 1 6 8 C 3 4 2 8 C 3 4 4 8 C 3 4 6 9 C 3 5 0 9 C 3 5 2 9 C 3 5 3 1 1 C 3 0 8 P .C N .C PBS 0.0 0.5 1.0 1.5 2.0 2.5 Hybridoma clone A b s a t 4 0 5 n m
Figure 9. ELISA for the binding of ascitic fluids containing anti-HSET mAbs to yeast displayed HSET fragment (Met1 - Ala100). HSET fragment displaying yeast cells (5 × 105
cells/well) were coated on 96-well plate and then 50-fold diluted ascitic fluids were treated. The
bound mAbs were detected by AP conjugated goat anti-mouse IgG. As negative controls,
irrelevant ascitic fluid (N.C) or PBS was used. Commercial anti-HSET mAb (from Abcam) was
used as a positive control. The first digit of the clone name stands for peptide antigen number.
24
Figure 10. SDS-PAGE of the purified anti-HSET mAbs. Anti-HSET mAbs collected from hybridoma cell culture supernatant were purified using protein G column. About 20 µg of each
protein was loaded on 12% acrylamiade gels and visualized with coomassie blue. In the
reducing condition, heavy chain and light chain protein are localized near 50 kDa and 25 kDa
25 1 C 2 7 2 1 C 2 7 4 1 C 2 7 5 2 C 2 7 9 2 C 2 8 0 2 C 2 8 1 3 C 2 8 8 3 C 2 8 9 3 C 2 9 0 3 C 2 9 2 4 C 2 9 5 4 C 2 9 8 5 C 3 0 3 5 C 3 0 4 6 C 4 0 4 6 C 4 0 6 6 C 4 0 7 7 C 3 1 5 7 C 3 1 6 8 C 3 4 2 8 C 3 4 4 8 C 3 4 6 9 C 3 5 0 9 C 3 5 2 9 C 3 5 3 1 0 C 3 5 8 1 0 C 3 6 2 1 1 C 3 8 0 a n ti His A b a n ti HS E T 1 -1 0 0 A b a n ti HS E T 6 2 5 -6 7 3 A b m o u s e I g G 0.0 0.5 1.0 1.5 2.0 2.5 Hybridoma clone A b s a t 4 0 5 n m
Figure 11. ELISA for the binding of the purified anti-HSET mAbs to whole HSET protein antigen. The anti-HSET mAbs (5 µg) purified from culture supernatants were added to the wells coated with whole HSET (Met1 - Lys673) protein (2.5 µg). The bound IgG mAbs were
detected using AP-conjugated anti-mouse Ab. Irrelevant polyclonal mouse IgG was used as a
negative control. As positive controles, commercial mAbs against N-terminus (1-50 aa) and
C-terminus (625-673 aa) of HSET, and His tag were used as positive controls. The first digit of the
clone name stands for peptide antigen number. Data represent the mean ± S.D. of triplicate
26
D. Comparison of binding activity between mAbs
We summarized the results of ELISA of Fig. 7, 8, 9, and 11 (Table 4). Clone 6C407 and
8C346 against Glu43 - Arg54 and Met1 - Lys12 of HSET, bound all kinds of antigen and were
expected to have high binding activity. Clone 2C280 and 2C281, these bound to whole HSET
protein, but not to yeast displayed protein. There are two clones that could bind peptide and
whole HSET protein but yeast displayed protein, clone 9C352 and 9C353 which have epitope
between Lys12 - Lys23. Because of different targeting region of clone 1C274 and 3C290
(Pro123 - Arg134 and Gln115 - Ala126, respectively), they were not available using Met1 -
Lys50 peptide and Met1 - Ala100 yeast displayed protein as antigens to perform ELISA.
Nevertheless, clone 1C274 and 3C290 showed comparatively high binding to whole HSET
protein. All of these clones were targeting N-terminus of HSET, not C-terminus. The reason
could be explained when amino acid sequence of human HSET and mouse KIFC1, were aligned.
Because C-terminus region of HSET and KIFC1 were almost similar sequence, it is thought that
27 Table 4. Summary of anti-HSET mAbs
MAbs in ascitic fluids Purified mAbs
Peptide (Met1 - Lys50)
Yeast displayed protein ( Met1 - Ala100) Whole protein (Met1 - Lys673) 1C272 NDa ND +b 1C274 ND ND +++++ 1C275 ND ND + 2C279 ND - - 2C280 ND - +++++ 2C281 ND + +++++ 3C288 ND ND + 3C289 ND ND ++ 3C290 ND ND +++ 3C292 ND ND + 4C295 - - - 4C298 - - +++ 5C303 ND ND + 5C304 ND ND ++ 6C404 - + + 6C406 - - +++ 6C407 +++++ ++ +++++ 7C315 - ++ + 7C316 - + + 8C342 - - + 8C344 - - + 8C346 +++++ +++ ++ 9C350 - + ++ 9C352 +++++ - +++++ 9C353 +++++ - +++++ 10C358 ND ND + 10C362 ND ND + 11C380 ND - ++ a Not determined b
The binding activity of mAbs was expressed as absorbance of samples
divided by negative control value (1-1.5: +, 1.5-2: ++, 2-2.5: +++, 2.5-3:
28
Figure 12. Sequence comparison between human HSET and mouse KIFC1. Human HSET and its mouse homolog, KIFC1, have higher homology of amino acid sequence at C-terminus
29 E. Sequence analysis of anti-HSET mAbs
Sequence of mAbs were analyzed for the generation or enginieering of recombinant Ab.
DNA sequences of all mAbs were analyzed regardless of their binding activities. It is because
that even mAbs with low binding activity against the prepared HSET could strongly bind to the
intracellularly expressed HSET. Variable region genes were amplified and read by the primers
that hybridize to immunoglobulin genes (Table 1). In case of clone 5C303 and 6C406, Vĸ genes
were not able to be identified because those genes were amplified by MKV2 primer which also
annealed to pseudo Vĸ gene from myeloma fusion partner cell. In case of clone 8C342, VH gene
was not amplified by any MHV primers for unknown reason.
Comparing the sequences of mAbs, mAbs that target same region of HSET had similar
variable region sequence (Table 5). Variable heavy chain genes (VH) of 3C288, 3C289, 3C290,
and 3C292 that target Gln115 - Ala126 (peptide 3) were 100% identical. In particular, 3C288
and 3C289 had 100% identical variable kappa chain (Vĸ). Clone 6C404 and 6C406 that target
Glu43 - Arg54 (peptide 6) had 100% identical VH sequence, although their Vĸ sequences could
not be compared. Clone 4C295 and 4C298 that recognize Ser33 - Asp44 (peptide 4) also had
30
Table 5. Amino acids sequence of variable regions in anti-HSET mAbs
31 Variable kappa chain
32
IV. DISCUSSION
It is well known that several cancer cells have amplified centrosomes compare with normal
cells. There were many attempts to target centrosome bundling mechanism as a development of
therapeutic molecules, including antifungal drug griseofulvin (Rebacz et al., 2007), kinesin
spindle protein inhibitor, CENP-E inhibitor (Huszar et al., 2009), and phenanthrene-derived
PARP inhibitor (Castiel et al., 2011). In recent studies, some small molecules that have been
developed to block the centrosome clustering function of HSET induced the death of cancer
cells, suggesting that HSET would be a promising target molecule for cancer therapy (Watts et
al., 2013; Wu et al., 2013).
Generally, comparing to small molecule inhibitors which can cause side effects from the
lack of specificity, mAbs are known to have high specificity against target antigen (Huang et al.,
2004). Moreover, the half-lives of mAbs are 4-fold longer than small molecule inhibitors
(Dancey and Sausville, 2003).
In this study, we successfully produced mAbs against N-terminus region of HSET
molecules using a hybridoma technique, with the expectation to inhibit the function of HSET in
the cancer cells in which function of HSET is indispensable for their cell division. HSET
sequences were analyzed by BLAST search to design antigen for immunization. Some regions
of HSET which show relatively hydrophilic and low homology with other proteins were
synthesized as 12 amino acid-length peptides. The advantage of short length antigens, instead of
immunization with full length protein, is that the production of anti-HSET mAbs which show
cross-reactivity with other kinesin could be avoided. Though several mAbs to peptide or whole
33
molecule through intracelluar expression of these mAbs should be confirmed by the method of
immunoprecipitation and immunofluorescence.
Binding activity of mAbs was different, depending on the types of antigen. It may be two
reasons as follows. First, the glycosylation pattern may affect the binding site of mAbs. HSET
fragment displayed on the yeast surface can be glycosylated in unpredicted regions of protein.
Different glycosylation pattern can lead to the change of protein conformation. Second, mAbs
that had been derived from the immunization with HSET peptide antigens could not recognize
the secondary/tertiary structure of intact HSET protein.
In fact, we tried simultaneously a hybridoma technique and a phage display technique to
obtain anti-HSET mAbs. Unfortunately, however, none of the mAbs was obtained from phage
display technique in which phage surface-displayed human Ab (single chain variable fragments)
library was panned against the peptide antigens (corresponding to Met1 - Ala100 and Gln625 -
Lys673 of HSET, respectively) as well as yeast surface-displayed HSET fragment (Met1 -
Ala100).
All most of IgG mAbs of 150 kDa cannot enter cells. To target intracellular molecules
using an Ab, internalization of Ab should be allowed. As a matter of fact, many researchers pay
attention to intracellular targeting Ab and a variety of attempts are being tried to penetrate Ab
into the cells (Kaiser et al., 2014). HSET is localized in nucleus and cytosol during interphase
and mitosis (Cai et al., 2009), respectively. It implies that anti-HSET Ab should enter across the
cellular membrane to target HSET molecule. Therefore, to inhibit HSET function by the
treatment of anti-HSET mAbs to cancer cells, the conversion of non-internalizing anti-HSET
34
V. CONCLUSION
We generated mouse mAbs against HSET protein involved in centrosome clustering
essential for division of the cancer cells with extra-centrosome, and analyzed the binding
35
REFERENCES
1. Aneja R, Vangapandu SN, Lopus M, Viswesarappa VG, Dhiman N, Verma A, Chandra
R, Panda D, Joshi HC: Synthesis of microtubule-interfering halogenated noscapine
analogs that perturb mitosis in cancer cells followed by cell death. Biochem Pharmacol
72: 415-426, 2006
2. Basto R, Brunk K, Vinadogrova T, Peel N, Franz A, Khodjakov A, Raff JW:
Centrosome amplification can initiate tumorigenesis in flies. Cell 133: 1032-1042, 2008
3. Boveri T: Concerning the origin of malignant tumours by Theodor Boveri. Translated
and annotated by Henry Harris. J Cell Sci 121 Suppl 1: 1-84, 2008
4. Cai S, Weaver LN, Ems-McClung SC, Walczak CE: Kinesin-14 family proteins
HSET/XCTK2 control spindle length by cross-linking and sliding microtubules. Mol
Biol Cell 20: 1348-1359, 2009
5. Castiel A, Visochek L, Mittelman L, Dantzer F, Izraeli S, Cohen-Armon M: A
phenanthrene derived PARP inhibitor is an extra-centrosomes de-clustering agent
36
6. Chames P, Van Regenmortel M, Weiss E, Baty D: Therapeutic antibodies: successes,
limitations and hopes for the future. Br J Pharmacol 157: 220-233, 2009
7. Dancey J, Sausville EA: Issues and progress with protein kinase inhibitors for cancer
treatment. Nat Rev Drug Discov 2: 296-313, 2003
8. Doxsey S: Re-evaluating centrosome function. Nat Rev Mol Cell Biol 2: 688-698, 2001
9. Fielding AB, Dobreva I, Dedhar S: Beyond focal adhesions: integrin-linked kinase
associates with tubulin and regulates mitotic spindle organization. Cell Cycle 7:
1899-1906, 2008
10. Godinho SA, Kwon M, Pellman D: Centrosomes and cancer: how cancer cells divide
with too many centrosomes. Cancer Metastasis Rev 28: 85-98, 2009
11. Goldenberg DM, Sharkey RM: Using antibodies to target cancer therapeutics. Expert
Opin Biol Ther 12: 1173-1190, 2012
12. Gordon MB, Howard L, Compton DA: Chromosome movement in mitosis requires
microtubule anchorage at spindle poles. J Cell Biol 152: 425-434, 2001
13. Hirokawa N, Takemura R: Molecular motors and mechanisms of directional transport in
37
14. Huang S, Armstrong EA, Benavente S, Chinnaiyan P, Harari PM: Dual-agent molecular
targeting of the epidermal growth factor receptor (EGFR): combining anti-EGFR
antibody with tyrosine kinase inhibitor. Cancer Res 64: 5355-5362, 2004
15. Huszar D, Theoclitou ME, Skolnik J, Herbst R: Kinesin motor proteins as targets for
cancer therapy. Cancer Metastasis Rev 28: 197-208, 2009
16. Kaiser PD, Maier J, Trankle B, Emele F, Rothbauer U: Recent progress in generating
intracellular functional antibody fragments to target and trace cellular components in
living cells. Biochim Biophys Acta, 2014
17. Karna P, Rida PC, Pannu V, Gupta KK, Dalton WB, Joshi H, Yang VW, Zhou J, Aneja
R: A novel microtubule-modulating noscapinoid triggers apoptosis by inducing spindle
multipolarity via centrosome amplification and declustering. Cell Death Differ 18:
632-644, 2011
18. Kleylein-Sohn J, Pollinger B, Ohmer M, Hofmann F, Nigg EA, Hemmings BA,
Wartmann M: Acentrosomal spindle organization renders cancer cells dependent on the
kinesin HSET. J Cell Sci 125: 5391-5402, 2012
19. Kwon M, Godinho SA, Chandhok NS, Ganem NJ, Azioune A, Thery M, Pellman D:
Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes.
38
20. Manandhar G, Schatten H, Sutovsky P: Centrosome reduction during gametogenesis
and its significance. Biol Reprod 72: 2-13, 2005
21. Ogden A, Rida PC, Aneja R: Let's huddle to prevent a muddle: centrosome declustering
as an attractive anticancer strategy. Cell Death Differ 19: 1255-1267, 2012
22. Quintyne NJ, Reing JE, Hoffelder DR, Gollin SM, Saunders WS: Spindle multipolarity
is prevented by centrosomal clustering. Science 307: 127-129, 2005
23. Raab MS, Breitkreutz I, Anderhub S, Ronnest MH, Leber B, Larsen TO, Weiz L,
Konotop G, Hayden PJ, Podar K, Fruehauf J, Nissen F, Mier W, Haberkorn U, Ho AD,
Goldschmidt H, Anderson KC, Clausen MH, Kramer A: GF-15, a novel inhibitor of
centrosomal clustering, suppresses tumor cell growth in vitro and in vivo. Cancer Res
72: 5374-5385, 2012
24. Rebacz B, Larsen TO, Clausen MH, Ronnest MH, Loffler H, Ho AD, Kramer A:
Identification of griseofulvin as an inhibitor of centrosomal clustering in a
phenotype-based screen. Cancer Res 67: 6342-6350, 2007
25. Ring D, Hubble R, Kirschner M: Mitosis in a cell with multiple centrioles. J Cell Biol
94: 549-556, 1982
26. Salisbury JL, Lingle WL, White RA, Cordes LE, Barrett S: Microtubule nucleating
39
27. Sharp DJ, Rogers GC, Scholey JM: Microtubule motors in mitosis. Nature 407: 41-47,
2000
28. Watts CA, Richards FM, Bender A, Bond PJ, Korb O, Kern O, Riddick M, Owen P,
Myers RM, Raff J, Gergely F, Jodrell DI, Ley SV: Design, synthesis, and biological
evaluation of an allosteric inhibitor of HSET that targets cancer cells with
supernumerary centrosomes. Chem Biol 20: 1399-1410, 2013
29. Williams D, Matthews D, Jones T: Humanising Antibodies by CDR Grafting. In
Antibody Engineering (eds. Kontermann R, Dübel S), Springer Berlin Heidelberg,
pp.319-339, 2010
30. Wu J, Mikule K, Wang W, Su N, Petteruti P, Gharahdaghi F, Code E, Zhu X, Jacques K,
Lai Z, Yang B, Lamb ML, Chuaqui C, Keen N, Chen H: Discovery and mechanistic
study of a small molecule inhibitor for motor protein KIFC1. ACS Chem Biol 8:
40 - 국문요약 -
HSET 에 대한 재조합 단클론 항체 생산
아주대학교 대학원 의생명과학과 서 영 실 (지도교수 : 권 명 희)HSET (Human spleen Embryo Testis)은 키네신 14군에 속한 단백질로서 세포 분열 시 중심체의 군집 (centrosome clustering)에 필수적인 역할을 한다. 세 개 이상의 중심체를 가진 암세포의 경우에는 다극성으로 분열 (multi-polar division) 시, 유전체 불안정성과 세포 사멸을 일으킬 수 있다. 이러한 암세포는 다극성 분열을 회피하기 위해 HSET을 필요로 하는데, 이는 HSET에 의하여 중심체가 군집화 되면 다극성 분열이 억제되고 정상적인 염색체의 분리가 가능해지기 때문이다. 본 연구에서는 암세포에서 HSET의 기능을 저해하기 위한 첫 번째 단계로서, 하이브리도마 기법을 이용하여 생쥐로부터 항-HSET 단클론 항체를 제작하여 항원에 대한 친화성을 분석하였다. 생쥐에 면역하기 위한 항원으로는 다른 키네신과의 상동성이 낮은 HSET의 N 말단 부위 (1-134번째 아미노산)와 C 말단 부위 (626-672번째 아미노산)에 해당하는 12개 아미노산으로 구성된 펩타이드를 사용하였다. 총 28개의 항체를 얻었으며 모두 HSET의 N 말단에 특이적이었다. 효소면역측정법을 사용하여 생쥐의 복수 속에 존재하는 항체와 효모
41 표면에 발현시킨 HSET과의 결합을 확인했을 때 클론 8C346이 높은 결합력을 보였고, 하이브리도마 배양액으로부터 정제된 항체의 HSET 단백질과의 결합력은 클론 1C274, 2C280, 2C281, 6C407, 9C352, 9C353에서 상대적으로 높게 나타났다. 이들 항체가 HSET의 기능을 억제하여 암세포 특이적인 사멸을 유도할 것으로 기대된다. 핵심어 : HSET, 중심체 군집, 항-HSET 항체, 하이브리도마, 암