의학
의학
의학
의학 박사학위
박사학위
박사학위
박사학위 논문
논문
논문
논문
Expression and Functional Analysis of
Hepatitis B Virus DNA Polymerase
아
아
아
아 주
주
주 대
주
대
대 학
대
학
학
학 교
교 대
교
교
대
대
대 학
학
학
학 원
원
원
원
의
의
의
의 학
학
학 과
학
과
과
과
박
박
박
박 길
길
길 순
길
순
순
순
Expression and Functional Analysis of
Hepatitis B Virus DNA Polymerase
by
Gil-Soon Park
A Dissertation Submitted to The Graduate School of Ajou University
in Partial Fulfillment of the Requirements for the Degree of
DOCTOR OF PHILOSOPHY
Supervised by
Kyongmin Kim, Ph.D.
Department of Medical Sciences
The Graduate School, Ajou University
박길순의
박길순의
박길순의
박길순의 의학
의학
의학
의학 박사학위
박사학위
박사학위
박사학위 논문을
논문을
논문을 인준함
논문을
인준함
인준함
인준함....
심사위원장
심사위원장
심사위원장
심사위원장 신
신
신
신
호
호
호
호
준
준
준
준
인
인
인
인
심 사 위 원
심 사 위 원
심 사 위 원
심 사 위 원 조
조
조
조 성
성
성
성 원
원
원 인
원
인
인
인
심 사 위 원
심 사 위 원
심 사 위 원
심 사 위 원 안
안
안
안 명
명
명
명 희
희
희 인
희
인
인
인
심 사 위 원
심 사 위 원
심 사 위 원
심 사 위 원 조
조
조
조 혜
혜
혜
혜 성
성
성 인
성
인
인
인
심 사 위 원
심 사 위 원
심 사 위 원
심 사 위 원 김
김
김
김 경
경
경
경 민
민
민 인
민
인
인
인
아
아
아
아 주
주
주 대
주
대
대 학
대
학
학
학 교
교 대
교
교
대
대
대 학
학
학
학 원
원
원
원
2005
2005
2005
2005 년
년
년
년 6
6
6 월
6
월
월
월 22
22
22 일
22
일
일
일
Expression and Functional Analysis of
Hepatitis B Virus DNA Polymerase
PART I
Expression of chimeric DNA polymerase of
human and duck hepatitis B virus
i
- ABSTRACT -
Expression and Functional Analysis of
Hepatitis B Virus DNA Polymerase
Part I
Expression of chimeric DNA polymerase of human and duck
hepatitis B virus
Hepatitis B Virus (HBV) DNA polymerase is a multifunctional protein with four domains, terminal protein (TP), spacer, reverse transcriptase (RT), and RNaseH. Since HBV DNA polymerase is very unstable and rapidly degradable, it has been very difficult to study the biochemical aspect of HBV DNA polymerase. In the present study, several chimeras of DNA polymerase by substituting DHBV sequence in the HBV DNA polymerase were constructed to have insights for the nature of HBV DNA polymerase. Also rabbit polyclonal antiserum specific for each domain of HBV DNA polymerase was produced. RNAs of chimeric DNA polymerases, and 2.4 and 2.1 Kbp of surface RNAs were produced by all chimeras but the expression proteins of chimeric polymerases were not detected. The distribution of HBV polyemerase was exclusively localized in cytoplasm and little co-localized between HBV polymerase and the organelles markers such as endoplasmic reticulum, Golgi and peroxisome. When the several chimeric constructs and polymerase deficient
ii
mutant that provides pgRNA and HBV proteins except polymerase, were co-transfected to hepatoma cells, all chimeras were incompatible for replication. Interestingly core particle formation varied in chimeric DNA polymerase construct transfected cells. This result suggests that the primary sequence of chimeric DNA polymerase may affect the capability core particle formation or the level of pregenomic RNA.
Key Words: Hepatitis B Virus DNA polymerase, Chimera, replication, core particle formation
iii
- ABSTRACT -
Expression and Functional Analysis of
Hepatitis B Virus DNA Polymerase
PART II
Down-regulation of hepatitis B virus replication by splicing event
Various spliced HBV transcripts have been described in human liver tissues and in HBV-transfected hepatoma cell lines, but their functions are unknown. To analyze the expression and the function of HBV DNA polymerase, HBV DNA polymerase construct was tranfected into hepatoma cells and then several spliced HBV RNAs from expressed HBV DNA polymerase RNA were founded. Since the spliced RNA encoded new viral protein, the polymerase-surface (PS) fusion protein was analyzed in detail. By confocal microscopy with monoclonal antibody specific for surface antigen and polyclonal antiserum specific for TP domain of HBV DNA polymerase, the exclusive expression of PS fusion protein in perinuclear region was detected. Also, intracellular distribution of PS fusion protein was overlapped with nuclear pore complex and vimentin, which is known as intermediate filament providing a perinuclear stable core area, as well as endoplasmic reticulum. Also, HBV small antigen secretion was drastically inhibited even with a small amount of
iv
this construct. Since it is possible that PS fusion protein expression might be involved in HBV life cycle by modulating gene regulation through nuclear/cytoplasmic transport like Rev protein of HIV, the spliced RNA only expressing mutant that did not express PS protein to elucidate the modulatory effects of the spliced RNA or the PS fusion protein on HBV life cycle. Core particle formation was blocked by the expression of spliced construct. Also, when the PS fusion RNA expression was increased, HBV DNA synthesis was decreased gradually by endogenous polymerase assay and Southern blot analysis. These data indicate that PS fusion RNA down-regulates HBV DNA synthesis, which distinguishes the HBV large surface RNA with PS fusion RNA. Taken together, these data suggest that PS fusion RNA may modulate the HBV replication, which is different from the reported previously.
Key Words: splicing, polymerase-surface (PS) fusion protein, replication, core particle formation
v
TABLE OF CONTENTS
ABSTRACT --- i
TABLE OF CONTENTS--- v
LIST OF FIGURES ---ix
LIST OF TABLES ---xi
PART I І. INTRODUCTION --- 1
II. MATERIALS AND METHODS --- 6
A. Construction of prokaryotic expression vector for the expression of HBV DNA polymerase domain --- 6
B. Expression and purification of the each domain in E. coli --- 7
C. Construction of vector for chimeric DNA polymerase expression --- 7
D. Cell culture and transfection --- 18
E. Immunofluorescence assay --- 18
F. Isolation of core particles --- 19
G. Northern and Southern blotting --- 19
H. Endogenous polymerase assay (EPA) --- 20
I. Encapsidation assay --- 20
vi
III. RESULTS --- 22
A. Determination of substituted regions between HBV and DHBV DNA polymerase --- 22
B. HBV and DHBV chimeric DNA polymerase constructions --- 25
C. Generation of polyclonal antibody specific for each domain of HBV DNA polymerase expressed from E. coli --- 27
D. Chimeric DNA polymerase constructs cannot support the encapsidation and replication of HBV --- 29
E. Chimeric DNA polymerase RNA expression --- 32
F. HBV DNA polymerase expression --- 34
G. The Core particle formation in chimeric DNA polymerase constructs transfected HuH7 cells --- ---38
IV. DISCUSSION --- 41 V. CONCLUSION --- 46 REFERENCES --- 48 국문요약 --- 56 PART II I. INTRODUCTION --- 60
II. MATERIALS AND METHODS --- 64
vii
B. Cell culture and transfection --- 67
C. RT-PCR --- 67
D. Isolation of core particles --- 68
E. Immunoprecipitation and Western blotting --- 68
F. Immunofluorescence assay --- 69
G. Enzyme-linked immunosorbent assay (ELISA) --- 70
H. Glycosylation stage analysis --- 70
I. Northern and Southern blotting --- 70
J. Endogenous polymerase assay --- 71
K. RNase protection assay (RPA) --- 72
III. RESULTS --- 73
A. Detection of the spliced RNAs and the spliced product, polymerase -surface fusion protein, from HBV polymerase expressing cells --- 73
B. Various spliced RNAs were detected in HBV polymerase expressing cells --- 79
C. Distribution of the PS fusion protein was distinct from the middle and the small surface protein --- 82
D. The PS fusion protein was partially overlapped with nuclear pore complex and a major of the PS protein was extensively overlapped with intermediate filament --- 88
E. The expression of the PS fusion construct was drastically inhibited surface protein secretion --- 93
viii
F. HBV DNA synthesis and core particle assembly were reduced and
delayed by the expression of PS fusion construct --- 96
G. The expression of the PS construct modulated the HBV life cycle by down-regulating the pg RNA but not the surface mRNA --- 99
IV. DISCUSSION --- 103
V. CONCLUSION --- 108
REFERENCES --- 109
ix
LIST OF FIGURES
PART I
Fig. 1. Amino acid sequence alignment of HBV and DHBV DNA polymerase --- 23 Fig. 2. Schematic diagram of chimeric DNA polymerase constructs with substituted
DHBV polymerase sequence --- 26 Fig. 3. The bacterial expression and the purification of GST-fused HBV
polymerase domains--- 28 Fig. 4. Chimeric DNA polymerase constructs cannot support HBV replications --- 31 Fig. 5. HBV RNA expressions from chimeric DNA polymerase transfected
HuH7 cells --- 33 Fig. 6. HBV DNA polymerase expressions and cellular distribution from wild type
of HBV, HBV DNA polymerase, and chimeric DNA polymerase
constructs transfected HuH7 cells --- 37 Fig. 7. Core particle formation form P deficient mutant and chimeric
x
PART II
Fig. 1. Detection and characterization of a spliced HBV transcript and spliced
protein from various HBV polymerase construct transfected cells --- 77
Fig. 2. The detection of another spliced transcript, the determination of the splice sites and the estimation of expected protein size from the spliced RNA in HBV polymerase transfected cells --- 81
Fig. 3. Schematic representation of the various mutants used in this study --- 84
Fig. 4. Characterizations of the polymerase-surface RNA and polymerase -surface fusion protein --- 87
Fig. 5. Intracellular distribution of the PS protein --- 91
Fig. 6. Effect of the PS construct for surface protein secretion --- 95
Fig. 7. Effect of the PS construct on HBV replication --- 98
xi
LIST OF TABLES
PART I
1
I. INTRODUCTION
Hepatitis B Virus (HBV) is one of the major human pathogen that can cause lifelong infection, cirrhosis (scarring) of the liver, liver cancer, liver failure, and death. Of the 2 billion people who have been infected with the HBV, more than 350 million have chronic hepatitis. Although new infections are preventable through vaccination, the treatment of HBV infection with nucleoside analogs is limited by the emergence of drug-resistant viruses. Therefore, the understanding of the nature of HBV replication and their components is a prerequisite for the solution of the serious global health problem (Das et al., 2001).
Hepadnaviruses are a family of small, enveloped DNA-containing viruses that replicate via reverse transcription of an pregenome RNA (pgRNA) within cytoplasmic core particle composed of viral core protein (Ganem D, 2001). Although significant differences exist among various hepadnaviruses, they share high degree of hepatotropism followed by the same replication cycle and are nearly identical in genetic organization. HBV is the prototype of hepadnavirus and the related animals viruses of woodchucks (woodchuck hepatitis B virus) and ducks (duck hepatitis B virus) have become important model system due to the lack of suitable in vivo experimental systems.
Residing within the core particle is the compact 3.2-Kbp partially double-stranded DNA genome that exists in a relaxed circular configuration with the genetic information organized into overlapping open reading frames coding for the surface,
2
core, polymerase and X proteins (Kocak et al., 2000).
Hepadnavirus polymerase plays a central role in the viral life cycle as a structural component for the polymerase-pgRNA complex formation for encapsidation and acts as a primer for minus-strand DNA synthesis and an enzyme for DNA synthesis. Polymerase proteins are composed of four domains; the terminal protein (TP) involves in the priming of reverse transcription by forming phosphodiester bonds between the hydroxyl group of Tyr and the first nucleotide. (Weber et al., 1994; Zoulim and Seeger, 1994; Lanford et al., 1997). The spacer region is not essential for polymerase function. Reverse transcriptase (RT) and RNase H domains of HBV polymerase display homology with RT of retrovirus with highly conserved sequence motifs of RTs and RNase H(Loeb et al., 1991).
The hepadnavirus replication cycle starts when N-terminal domain of the large envelope protein (L protein) of the virus binds to the unidentified receptor of hepatocyte (Ganem D, 2001; Hourioux et al., 2002). Fusion of the viral envelope with a cellular membrane liberates the subviral core particle into the cell cytoplasm. Once in nucleus, core particle releases the partially double-stranded relaxed circular viral DNA. And then this DNA is repaired to a covalently closed circular episome as template for transcription via host RNA polymerase II (Yao et al., 2000). The viral mRNAs are exported to the cytoplasm through nuclear pore complex. PgRNA acts as mRNA translated for core protein and polymerase. After translation, encapsidation is followed, in which pgRNA is selectively packaged into viral core particles as a ribonucleoprotein (RNP) complex with viral polymerase. Hepadnavirus DNA
3
replication is initiated when TP of polymerase is covalently linked to newly synthesized nucleotide as a protein primer without a nucleic acid primer, at a bulge region of epsilon (ε), the encapsidation signal, resulting in the synthesis of short oligomer (Tavis et al., 1994; Wang et al., 1994; Nassal and Rieger, 1996). However, recent work had indicated that protein priming is not a prerequisite for initiation of DNA synthesis since short oligomer is synthesized without protein priming reaction (Kim et al., 2004). Even so this protein priming reaction should be crucial for translocation since priming deficient mutant cannot translocate the nascent DNA (Kim et al., 2004). The DNA-polymerase complex translocates from ε to direct repeat (DR) 1 at the 3' end of pgRNA, where the minus-stranded DNA synthesis resumes (Seeger et al., 1986; Seeger and Maragos, 1990, 1991; Condreay et al., 1992; Wang and Seeger, 1993; Tavis et al., 1994). Minus-strand DNA synthesis continues until the 5′ end of pgRNA, while RNA template is degraded by the RNase H activity of DNA polymerase except the short stretch of RNA. This short terminal RNA oligomer is translocated to DR2, where it serves as a primer for plus-strand DNA synthesis (Seeger and Maragos, 1989; Loeb et al., 1991; Staprans et al., 1991). Once plus-strand DNA synthesis has reached the 5′end of minus-strand DNA, a final translocation to the 3′end of minus-strand DNA occurs, resulting in partially double-stranded relaxed circular DNA genome, which is not covalently closed. The mature core particles with DNA are either transported back into the nucleus to maintain the pool of covalently closed circular DNA (cccDNA) or bud into the endoplasmic reticulum, where they pick up the surface glycoproteins. The virions are then
4
noncytopathically secreted from the cell (Yao et al., 2000).
Since HBV DNA polymerase is very unstable and rapidly degradable, HBV DNA polymerase is maintained at a very low concentration in cells. Therefore, it has been very difficult to study the biochemical aspects of HBV DNA polymerase. Heterologous expression of full-length polymerase capable of authentic ε-dependent priming has not yet been achieved. HBV polymerase produced from recombinant baculoviruses in insect cells exerts some DNA synthesis activity, but not ε-dependent (Lanford et al., 1995; Lanford et al., 1997). An enzymatically active DHBV polymerase fusion protein in Ty1 retrotransposon had been described and appeared stable but was active only inside the generatedvirus-like particles (Beck and Nassal, 2001). Also attempts to generate full-length priming-competent polymerase in bacteria have failed (Beck and Nassal, 2001).
Due to the limitation of the many approaches described above, new approach to elucidate the function of HBV polymerase and to identify the domain of HBV DNA polymerase critical for HBV replication is to employ studies with chimeric viruses. The requirements of retrovirus encapsidation was identified using the chimeric viruses (Berkowitz et al., 1995; Poon et al., 1998; Certo et al., 1999; Beasley and Hu, 2002; Mansky and Gajary, 2002). Phenotypic mixing of envelope proteins between two avian hepadnaviruses (Ishikawa and Ganem, 1995) and between HBV and woolly monkey HBV (Chouteau et al., 2001) had resulted in the species specificity conversion. Also in vitro expression of HBV genomes carrying woodchuck hepatitis virus pre-S1 sequences showed that this chimeric virus was
5
compatible with HBV DNA replication and virion formation (Hourioux et al., 2002). Heron hepatitis B virus (HHBV) and DHBV chimeric viruses have been successfully elucidated the interactions between cis-acting sequences that are important for the synthesis of plus-strand DNA (Mueller-Hill and Loeb, 2002). Evidence for functional interactions between viral components for pgRNA encapsidation was provided by duck and heron chimeric viruses (Ostrow and Loeb, 2004).
In this study, series of chimeric polymerases were constructed and expressed in hepatoma cells to map the essential motifs of HBV DNA polymerase for HBV replication. To analyze HBV DNA polymerase, polyclonal antiserum against each domains of DNA polymerase expressed from E. coli was generated from rabbit. For the constructions of chimeric polymerases of HBV and DHBV, amino acid sequence of DNA polymerase was aligned and then four domains of hepadnavirus polymerase were subdivided into N- and C-terminus. All of chimeric DNA polymerases were not compatible for encapsidation and DNA synthesis. Interestingly, chimeric polymerase expression influenced the core particle formation. Results suggest that RNA of chimeric DNA polymerase seemed to influence the formation of core by the primary sequence.
6
II. MATERIALS AND METHODS
A. Construction of prokaryotic expression vector for the expression of HBV DNA polymerase domain
HBV subtype adw R9, 1.3 length of the HBV genome (Blum et al., 1991), was used to generate the each domain of HBV DNA polymerase construct. The pGEX-KG, a prokaryotic expression vector was used (Guan and Dixon, 1991). TP domain was amplified by polymerase chain reaction (PCR) with sense primer HBV 29 that target nt 2309-2332 (5′-CGGGATCCGCCCCTATCTTATCAACACTTC-3′) containing BamHI site and antisense primer HBV 30 that target nt 2826- 2847 (5′- GCGGATCCTCTTGTTCCCAAGAATATGGTG-3′) of HBV genome. 540 bp of the PCR product was digested with BamHI, and cloned into the corresponding site of pET-15b (Invitrogen, Carlsbad, CA, USA). His tagged TP fragments were re-digested with NcoI and HindIII (New England Biolabs, Inc), and incubated with T4 DNA polymerase to make the blunt-ended DNA fragment and then cloned into SmaI site of pGEX-KG. For Spacer, RT and RNase H domains, HBV wt genome was digested with MscI and SspI, MscI and DraI, and MscI and XhoI (New England Biolabs, Beverly, MA, USA), respectively. Each of digested fragments were incubated with T4 DNA polymerase to make the blunt-ended DNA fragment and then cloned into SmaI site of pGEX-KG. PCR derived construct was sequenced to confirm the presence of specific HBV sequences and to ensure that no extraneous mutations were introduced during PCR.
7
B. Expression and purification of the each domain in E. coli
E. coli (MC1061 strain) were transformed with glutathione-S-transferase
(GST) fused HBV DNA polymerase domains in pGEX-KG expression vectors and grown at 37°C overnight in 10ml LB medium with 100 µg/ml of ampicillin. Overnight culture was put to 1L LB broth with 100 µg/ml of ampicillin and incubated at 37°C until the absorbance at 600 nm was reached 0.5- 0.8. After 3 hrs of 0.5mM IPTG induction at 37°C, bacteria were harvested by centrifugation at 10,000g for 5 min at 4°C, resuspended in lysis buffer (50mM Tris.Cl pH7.5, 0.5mM EDTA, 0.3M NaCl, proteanase inhibitor cocktail, 0.1% NP40) and sonicated in ice for 10 sec with short intervals for three times. After centrifugation of lysate at 10,000g for 15 min at 4°C, pellet was harvested, treated with DNaseI (3 µg/ml), and loaded onto SDS-PAGE gel. Fusion proteins were directly eluted from gel slice.
C. Construction of vector for chimeric DNA polymerase expression
Wild-type (wt) subtype adw R9 designated as a pPB and polymerase-deficient mutant of HBV designated as a P polymerase-deficient were constructed under the cytomegalovirus immediate early (IE) promoter (Kim et al., 2004). DHBV clone was kindly provided from Dr. W. Mason, Fox Chase Cancer Center, USA (NCBI accession number; K01834).
8
length of HBV polymerase gene, PCR was performed. In brief, HBV polymerase gene was amplified by PCR with the sense mutagenic primer HBV 23 that contained
HindIII site and Kozak sequence and the antisense primer HBV 21. The resulting 660
bp of PCR product was digested with HindIII and BstEII, and cloned into the corresponding restriction sites of pPB, yielding pHBV pol. To generate the DHBV polymerase gene under the CMV promoter, PCR and fragment ligation were performed. Part of DHBV polymerase gene was amplified by PCR with the sense mutagenic primer DHBV 1 that target nt 20-50 of DHBV genome (5′ -GGAATTCGCCACCATGCAAAAATTAACGACGAATCACTGGATAG-3′) and the antisense primer DHBV 2 that target nt 474-450 of DHBV genome (5′ -GTACAGCCCTTCATTTGATATAGTC-3′). The resulting 450 bp of PCR product was digested with EcoRI and BglII. The pcDNA3 vector was digested with same restriction enzyme and then two fragments from PCR and vector were ligated and redigested with BglII to obtain the ligated fragment. Full-length DHBV sequences were digested with BglII and EcoRI to get most of DHBV DNA polymerase, resulting 2.6 Kbp fragment. This 2.6 Kbp fragment was cloned into pcDNA3 vector that had been digested with same enzymes, resulting pDA. The BglII digested 1.3 Kbp fragment was cloned into the corresponding restriction sites of pDA, resulting pDHBV pol.
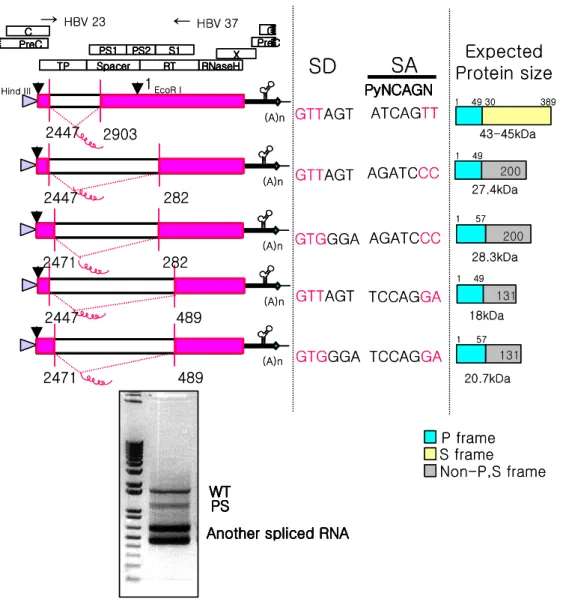
Primer sets used to construct chimeric DNA polymerases were shown in Table 1. The alignment of amino acid sequence of HBV DNA polymerase and DHBV DNA polymerase is shown in Fig. 1.
9
To generate pDTN construct under the CMV promoter that has N terminal region of DHBV TP, PCR was performed. The N-terminal region of DHBV TP domain was amplified by PCR with the antisense mutagenic primer HBV 44 and the sense primer PC 3, which binds to the CMV promoter region of pcDNA3. The HBV sequence to be fused with N-terminal region of DHBV TP was amplified by sense mutagenic primer HBV 45 and antisense primer HBV 21. The resulting 950 bp and 300 bp of PCR products were combined by fusion PCR. The fusion PCR product was digested with SnaBI and BstEII, and cloned into the corresponding restriction sites of wt HBV DNA polymerase, yielding pDTN. For pDTC that has C terminal region of DHBV TP, HBV was amplified with the antisense mutagenic primer HBV 47 and sense primer PC 3. The DHBV sequence to be fused with N-terminal region of HBV TP was amplified with sense mutagenic primer HBV 46 and antisense primer HBV 48 with BstEII site. The resulting 700 bp and 200 bp of PCR products were combined by fusion PCR. The 900 bp of fusion PCR product was digested with
BspEI and BstEII, and cloned into the corresponding restriction sites of wt HBV
DNA polymerase, yielding pDTC. For pDS that has spacer domain of DHBV, DHBV DNA polymerase was amplified with HBV 50, the antisense mutagenic primer and HBV 49, the sense mutagenic primer with BstEII. HBV 51, the sense mutagenic primer, and HBV 8, the antisense primer, was used to amplify the HBV sequence to be fused with spacer of DHBV. The resulting 500 bp and 600 bp of PCR products were combined by fusion PCR. The 1.1 Kbp of fusion PCR product was digested with XcmI and BstEII, and cloned into the corresponding restriction sites of wt HBV
10
DNA polymerase, yielding pDS. For pDRN that has the N terminal region of DHBV RT, DHBV DNA polymerase was amplified with HBV 56, the antisense mutagenic primer, and HBV 3, the sense primer. The HBV sequence to be fused with N-terminal region of DHBV RT was amplified by sense mutagenic primer HBV 57 and antisense mutagenic primer HBV 58. The other HBV sequence to be fused with N-terminal region of DHBV RT was amplified by sense mutagenic primer HBV 59 and antisense primer HBV 19. The resulting 170 bp, 400 bp and 1 Kbp of PCR products were combined for fusion PCR. The resulting 550 bp of the second fusion PCR product was combined with 1 Kbp first PCR product by third PCR. The resulting 1.6 Kbp PCR fragment was digested with EcoRI and EcoRV, and cloned into the corresponding restriction sites of wt HBV DNA polymerase, yielding pDRN. For pDRC that has the C-terminal region of DHBV RT, DHBV DNA polymerase was amplified with the antisense mutagenic primer HBV 60 and sense primer HBV 4. The DHBV sequence to be fused with C-terminal region of DHBV RTwas amplified by sense mutagenic primer HBV 61 and antisense mutagenic primer HBV 63. The other HBV sequence to be fused with C-terminal region of DHBV RT was amplified by sense mutagenic primer HBV 63 and antisense primer HBV 19. The resulting 380 bp, 450 bp and 640 bp of PCR products were combined by fusion PCR. The resulting 800 bp of second fusion PCR product was combined with 640 bp of first PCR products. The resulting 1.5 Kbp of third PCR fragment was digested with XcmI and
SacII, and cloned into the corresponding restriction sites of wt HBV DNA
11
RNase H, DHBV DNA polymerase was amplified with the antisense mutagenic primer HBV 64 and sense primer HBV 4. The DHBV sequence to be fused with N-terminal region of DHBV RNase H, DHBV sequence was amplified with sense mutagenic primer HBV 65 and antisense mutagenic primer HBV 66. The other HBV sequence to be fused with N-terminal region of DHBV RNase H, HBV sequence was amplified by sense mutagenic primer HBV 67 and antisense primer PC 5 that binds to the upstream of SP6 promoter region of pcDNA3. The resulting 800 bp, 300 bp and 500 bp of PCR products were combined by fusion PCR. The resulting 800 bp of second fusion PCR product was combined with 800 bp of first PCR product. The resulting 1.6 Kbp of third PCR fragment was digested with EcoRV and NotI, and cloned into the corresponding restriction sites of wt HBV DNA polymerase, yielding pDHN. For pDH1Cdε that has C-terminal 35 amino acids of DHBV RNase H, DHBV DNA polymerase was amplified with sense mutagenic primer HBV 52 and antisense primer PC 5 that binds to upstream of SP6 promoter of pcDNA3. The HBV sequence to be fused with C-terminal 35 amino acids of DHBV RNase H was amplified with sense primer HBV 18 and antisense mutagenic primer HBV 53. The resulting 110 bp and 670 bp of PCR products were combined by fusion PCR. 750 bp of fusion PCR product was digested with SacII and NotI, and cloned into the corresponding restriction sites of wt HBV DNA polymerase, yielding pDH1Cdε. For pDH2Cdε that has C-terminal 35 amino acids of DHBV RNase H, DHBV DNA polymerase was amplified with the sense mutagenic primer HBV 54 and sense primer PC 5. The HBV sequence to be fused with C-terminal 35 amino acids of
12
DHBV RNase H was amplified by sense primer HBV 18 and antisense mutagenic primer HBV 55. The resulting 160 bp and 670 bp of PCR products were combined by fusion PCR. The 800 bp of fusion PCR product was digested with SacII and NotI, and cloned into the corresponding restriction sites of wt HBV DNA polymerase, yielding pDH2Cdε. To replace the epsilon region of DHBV in DH1Cdε and pDH2Cdε to HBV, epsilon part of HBV from pPB was amplified with sense mutagenic primer HBV 72 and antisense primer PC 5. pDH1Cdε and pDH2Cdε were amplified with sense primer HBV 18 and antisense mutagenic primer HBV 73, respectively. The combined resulting 600 bp and 650 bp products were digested with
SacII and NotI, and cloned into the corresponding restriction sites of pDH1Cdε and pDH2Cdε constructs, yielding pDH1C and pDH2C construct, respectively. The pDH1Cdε and pDH1C constructs or the pDH2Cdε and pDH2C constructs have C-terminal 35 amino acid of DHBV RNase H region, but the joining site with HBV RNase H are different each other (Fig. 1). All constructs were sequenced to confirm the presence of specific mutations and to ensure that no extraneous mutations were introduced during PCR.
13
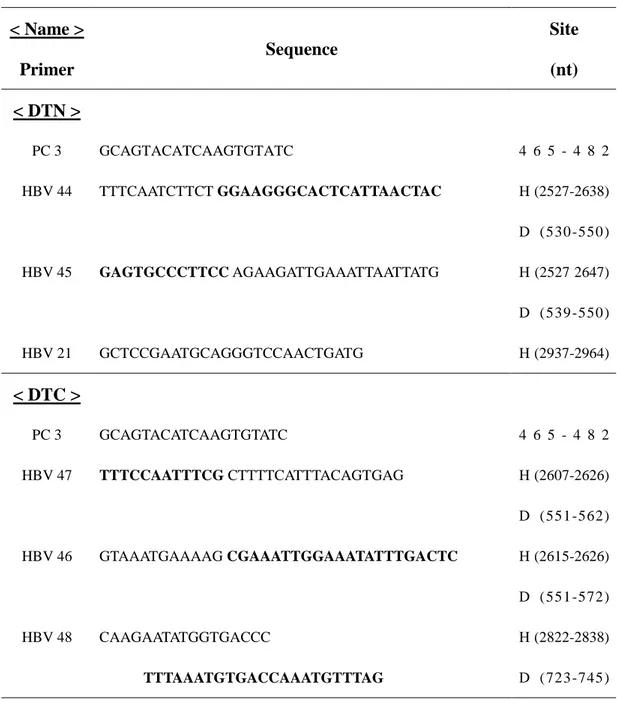
Table 1. Primers used for chimeric DNA polymerase construction. For chimeric
DNA polymerases construction, nucleotide positions of primers are given starting on the first dA residue of the EcoRI site of adw R9 wt genome and on the first dC residue of the EcoRI site of DHBV genome (NCBI accession number; K01834).
< Name > Primer Sequence Site (nt) < DTN > PC 3 HBV 44 HBV 45 HBV 21 GCAGTACATCAAGTGTATC TTTCAATCTTCT GGAAGGGCACTCATTAACTAC GAGTGCCCTTCC AGAAGATTGAAATTAATTATG GCTCCGAATGCAGGGTCCAACTGATG 4 6 5 - 4 8 2 H (2527-2638) D (530 -550 ) H (2527 2647) D (539 -550 ) H (2937-2964) < DTC > PC 3 HBV 47 HBV 46 HBV 48 GCAGTACATCAAGTGTATC TTTCCAATTTCG CTTTTCATTTACAGTGAG GTAAATGAAAAG CGAAATTGGAAATATTTGACTC CAAGAATATGGTGACCC TTTAAATGTGACCAAATGTTTAG 4 6 5 - 4 8 2 H (2607-2626) D (551 -562 ) H (2615-2626) D (551 -572 ) H (2822-2838) D (723 -745 )
14 (continued) < Name > Primer Sequence Site (nt) < DRN > HBV 3 HBV 56 HBV 57 HBV 58 HBV 59 HBV 19 CCACCTCT AAGAGACAGTC TTTTTATCACT GCGGAGATTGACGAGATGTG GTCAATCTCCGC AGTGATAAAAACTCCCCCCTTG CAGTGGGGGAA GCGACGAGAGATTTCGGATC CTCTCGTCGC TTCCCCCACTGTTTGGCTTTC GTGCGCAGACCAATTTATGC H (3183-3201) H (11 0 -1 31 ) D (1252-1263) H (120 -132 ) D (1253-1274) H (714 -724 ) D (1659-1678) H (714 -735 ) D (1669-1678) H (1789-1808)
15 (continued) < Name > Primer Sequence Site (nt) < DRC > HBV 4 HBV 60 HBV 61 HBV 62 HBV 63 HBV 19 CCAATCACTCACCAACCTCC CAAACGTTAA AGCCCTACGAACCACTGAAC TCGTAGGGCT TTTAACGTTTGGACTTTCAC CACAGACCAGG TGGCTTTATTCTTAATTTAC GAATAAAGCCA CCTGGTCTGTGCCAAGTGTTTG GTGCGCAGACCAATTTATGC H (328-3470) H (694 -713 ) D (1680-1689) H (704 -714 ) D (1679-1698) H (1167-1177) D (2112-2131) H (1167-1188) D (2121-2131) H (1789-1808)
16 (continued) < Name > Primer Sequence Site (nt) < DHN > HBV 5 HBV 64 HBV 65 HBV 66 HBV 67 PC 5 GCTTTCAGCTATATGGATG CAGAGGACTT CCGTTGCCGAGCAACGGGGT CTCGGCAACGG AAGTCCTCTGTACCTTTGC CAAGCGGCC CCTGGATGGGCCGTCAGCAGGA GCCCATCCAGG GGCCGCTTGGGACTCTCTCG GCATTTAGGTGACACTGTAG H (729 -747 ) H (1147-1166) D (2132-2141) H (1156-1166) D (2132-2150) H (1470-1478) D (2425-2446) H (1470-1489) D (2436-2446) 9 9 9 - 1 0 1 8
17 (continued) < Name > Primer Sequence Site (nt) <DH1Cdεεεε > HBV 18 HBV 53 HBV 52 PC 5 GTTTCCATGGCTGCTAGGTTG GAGGTTTGTG CCGAGAGGGGTCGTCCGCG CCCCTCTCGG CACAAACCTCCTGATTGGAC GCATTTAGGTGACACTGTAG H (1370-1390) H (1451-1469) D (2447-2456) H (1460-1469) D (2447-2466) 9 9 9 - 1 0 1 8 < DH2Cdεεεε > HBV 55 HBV 54 GAGGTTTGTG CGTGGTCGGCTGGAACGGCA GCCGACCACG CACAAACCTCCTGATTGGAC H (1504-1523) D (2447-2456) H (1514-1523) D (2447-2466) < DH1C, DH2C > HBV 73 HBV 72 GATGGGCGT TTAAGTTCCACATAGCCTATG GTGGAACTTAA ACGCCCATCAGATCCTGC H (1626-1639) D (2510-2530) H (1626-1643) D (2520-2530)
18
D. Cell culture and transfection
HuH7 hepatoma cell lines were used for the transfection of wt HBV DNA polymerase and chimeric DNA polymerase constructs. HuH7 cells were maintained in DMEM (Invitrogen, Carlsbad, CA, USA) supplemented with streptomycin and penicillin, and 10% fetal bovine serum (Invitrogen, Carlsbad, CA, USA). 2 µg of wt HBV DNA polymerase and chimeric polymerase constructs were transfected into HuH7 cells using lipofectamine (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions.
E. Immunofluorescence assay
Cells were seeded onto cover slip, incubation for 24-48 hrs, washed with phosphate-buffered saline (PBS), fixed in 4% paraformaldehyde for 10min, and then permeabilized in 0.15% Triton X-100 in PBS for 5 min. Samples were incubated with 3% bovine serum albumin (BSA) for 1 hr and then with the first antibody. Anti-calnexin antibody (Abcam Limited, Cambridgeshire, UK) was used to detect the distribution of ER. Anti-LMAN1 polyclonal IgY antibody (GenWay Biotech, Inc, San Diego, CA, USA) and anti-58K Golgi protein antibody (Abcam Limited, Cambridgeshire, UK) were used to detect the distribution of ER-Golgi intermediate compartment and Golgi apparatus. Peroxisome was detected using the anti-catalase antibody (LabFrontier, Seoul, Korea). The first antibodies in the 3% bovine serum
19
albumin were incubated at room temperature for 1 hr. Cells were incubated with anti-mouse IgG or anti-rabbit IgG coupled rhodamine or fluorescein isothiocynate dye for 1 hr. Cover slips were applied to the slides in mounting medium (VECTASHIELD, Vector Laboratories) and the cells were examined with a confocal laser scanning microscope (LSM510, Zeiss).
F. Isolation of core particles
Three days after transfection, HuH7 cells were lysed in 1 ml TNE (10mM Tris-HCl [pH 8.0], 50mM NaCl, 1mM EDTA)-1% Noniodet P-40. Then their nuclei were removed by centrifugation. The clarified lysate was adjusted with 10mM MgCl2
and 8mM CaCl2 solution, and incubated for 30 min at 37°C with 10 U DNase I
(Roche Applied Science, Indianapolis, IN, USA) and 30 U micrococcal nuclease (Calbiochem, Darmstadt, Germany). Cytoplasmic core particles were precipitated with 6.5% polyethylene glycol.
G. Northern and Southern blotting
Total RNA was extracted from cells using RNAzol B (TEL-TEST INC, Friendswood, Texas). In brief, cells were lysed with 0.2 ㎖ of RNAzol B per 1×106 cells, then 20 ㎕ chloroform was added to lysate. After vigorous shaking, the cell lysate was incubated on ice for 5 min and centrifuged at 12,000 g for 15 min at 4℃.
20
RNA in the aqueous phase that transferred to fresh tube was precipitated by addition of equal volume of isopropanol. After centrifugation, the RNA pellet was dried and then dissolved in RNase-free distilled water. 5 µg of total RNA was denatured and electrophoresed on a 1% agarose gel containing formaldehyde and blotted onto a nylon membrane. RNA on the membranes was hybridized to a [32P]-labeled random-primed probe specific for the HBV sequence. To analyze HBV DNA synthesis by Southern blotting, core DNA was extracted, separated by agarose gel electrophoresis, and hybridized to a [32P]-labeled random-priming HBV specific for the HBV sequence.
H. Endogenous polymerase Assay (EPA)
Isolated core particles were incubated at 37°C for 8-12 hrs with EPA reaction buffer (50mM Tris-HCl [pH 7.5], 75mM NH4Cl, 1mM EDTA, 25mM MgCl2, 0.1%
β-meracaptoethanol, 0.5% Noniodet P-40) supplemented with 0.5mM each of dCTP, dGTP and dTTP, and 10µCi α-32P-dATP (3000 Ci/mmol, Amersham Biosciences, Piscataway, NJ, USA). 32P-labeled reaction mixtures were electrophoresed on a 1% agarose gel and subjected to autoradiography. 32P –labeled DNA was extracted following the EPA reaction and separated by 1% agarose gel electrophoresis.
21
The pellet of isolated core particles was dissolved in 15 µl of Tris-acetate EDTA buffer and electrophoresed on a 1% native agarose gel. Core particles were transferred to nylon membrane. Transferred core particles were denatured with 0.2N NaOH in situ and neutralized. Nucleic acids from disrupted core particles were hybridized to a [ 32P]-labeled random- primed probe specific for the HBV sequence.
J. Core particle Western blotting
.
The pellet of isolated core particles was dissolved in 15 µl of Tris-Acetate EDTA buffer and electrophoresed on a 1% native agarose gel. Core particles were transferred to polyvinylidene fluoride (Schleichr & Schuell, Keene, NH, USA) membranes as described for Northern blot analysis. Immunoblotting was performed using a rabbit anti-hepatitis B virus core antigen (DakoCytomation Denmark A/S). Horseradish peroxidase-conjugated anti-rabbit secondary antibody and enhanced chemical luminescence (Amersham Biosciences, Piscataway, NJ, USA) were employed to visualize HBV core particles.
22
III. RESULTS
A. Determination of substituted regions between HBV and DHBV DNA polymerase
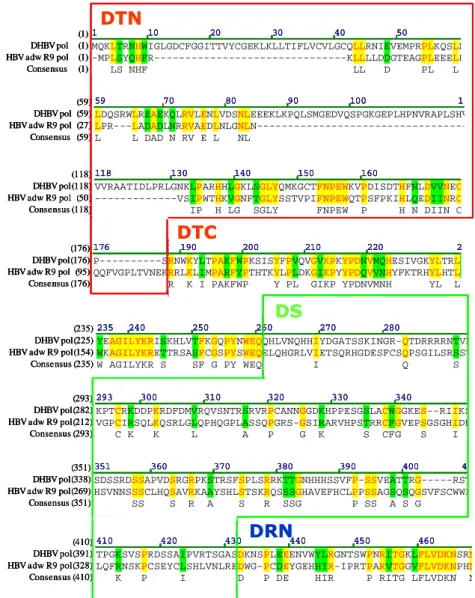
Amino acid sequences between HBV and DHBV DNA polymerase were aligned to determine which domain would be exchanged to identify the functional domains of HBV DNA polymerase (Fig. 1). To map essential domains or motifs of HBV DNA polymerase for HBV replication, chimeric DNA polymerase of HBV and DHBV were constructed in this study (Fig. 2). DHBV DNA polymerase share 28% identity with HBV DNA polymerase and therefore there is very little primary sequence identities between the two viral polymerases. Identical, weakly similar or non-similar sequences between two enzymes are presented as yellow, green and black, respectively (Fig. 1). The subdivided N- and C-terminus of each domain of hepadnavirus polymerase are presented as boxes in Fig. 1.
23
Fig. 1. Amino acid sequence alignment of HBV and DHBV DNA polymerase.
Identical, weakly similar or non-similar sequences between two enzymes are shown as yellow, green, and black, respectively. To construct chimeric DNA polymerase, each domain of hepadnavirus DNA polymerase are subdivided to N- and C-terminus and presented as various colored boxes.
1 10 20 30 40 50 61
(1)
MQKLTRNHWIGLGDCFGGITTVYCGEKLKLLTIFLVCVLGCQLLRNIEVEMPRPLKQSLD
DHBV pol (1)
-MPLSYQHFR---KLLLLDDGTEAGPLEEELP
HBV adw R9 pol (1) LS NHF LL D PL L Consensus (1) 59 70 80 90 100 119 (59) LDQSRWLREAEKQLRVLENLVDSNLEEEKLKPQLSMGEDVQSPGKGEPLHPNVRAPLSHV DHBV pol (59) LPR---LADADLNRRVAEDLNLGNL N---HBV adw R9 pol (27) L L DAD N RV E L NL Consensus (59) 118 130 140 150 160 (118)
VVRAATIDLPRLGNKLPARHHLGKLSGLYQMKGCTFNPEWKVPDISDTHFNLDVVNEC
DHBV pol(118)
---VSIPWTHKVGNFTGLYSSTVPIFNPEWQTPSFPKIHLQEDIINRC
HBV adw R9 pol (50)
IP H LG SGLY FNPEW P H N DIIN C
Consensus (118)
176 190 200 210 220 236
(176)
P---SRNWKYLTPAKFWPKSISYFPVQVGVKPKYPDNVMQHESIVGKYLTRLY
DHBV pol(176)
QQFVGPLTVNEKRRLKLIMPARFYPTHTKYLPLDKGIKPYYPDQVVNHYFKTRHYLHTLW
HBV adw R9 pol (95)
R K I PAKFWP Y PL GIKP YPDNVMNH YL LW
Consensus (176)
235 240 250 260 270 280
(235)
YEAGILYKRISKHLVTFKGQPYNWEQQHLVNQHHIYDGATSSKINGR-QTDRRRRNTVK
DHBV pol(225)
WKAGILYKRETTRSASFCGSPYSWEQELQHGRLVIETSQRHGDESFCSQPSGILSRSSV
HBV adw R9 pol(154)
W AGILYKR S SF G PY WEQ I Q S
Consensus (235)
293 300 310 320 330 340
(293)
KPTCRKDDPKRDFDMVRQVSNTRSRVRPCANNGGDKHPPESGSLACWGGKES--RIIKS
DHBV pol(282)
VGPCIRSQLKQSRLGLQPHQGPLASSQPGRS-GSIRARVHPSTRRCFGVEPSGSGHIDH
HBV adw R9 pol(212) C K K L A P G K S CFG S I Consensus (293) 351 360 370 380 390 400 411 (351) SDSSRDSSAPVDSRGRPKSTRSFSPLSRRKTTGNHHHSSVFP-SSVEATTRG---RST DHBV pol(338)
HSVNNSSSCLHQSAVRKAAYSHLSTSKRQSSSGHAVEFHCLPPSSAGSQSQGSVFSCWWL
HBV adw R9 pol(269) SS S R A S R SSG P SS A S G Consensus (351) 410 420 430 440 450 460 (410) TPGKSVSPRDSSAIPVRTSGASDKNSPLEEENVWYLRGNTSWPNRITGKLFLVDKNSRN DHBV pol(391)
LQFRNSKPCSEYCLSHLVNLREDWG-PCDEYGEHHIR-IPRTPARVTGGVFLVDKNPHN
HBV adw R9 pol(328) K P I D P DE HIR P RITG LFLVDKN N Consensus (410) DTN DTC DS DRN 1 10 20 30 40 50 61 (1) MQKLTRNHWIGLGDCFGGITTVYCGEKLKLLTIFLVCVLGCQLLRNIEVEMPRPLKQSLD DHBV pol (1)
-MPLSYQHFR---KLLLLDDGTEAGPLEEELP
HBV adw R9 pol (1) LS NHF LL D PL L Consensus (1) 59 70 80 90 100 119 (59) LDQSRWLREAEKQLRVLENLVDSNLEEEKLKPQLSMGEDVQSPGKGEPLHPNVRAPLSHV DHBV pol (59) LPR---LADADLNRRVAEDLNLGNL N---HBV adw R9 pol (27) L L DAD N RV E L NL Consensus (59) 118 130 140 150 160 (118)
VVRAATIDLPRLGNKLPARHHLGKLSGLYQMKGCTFNPEWKVPDISDTHFNLDVVNEC
DHBV pol(118)
---VSIPWTHKVGNFTGLYSSTVPIFNPEWQTPSFPKIHLQEDIINRC
HBV adw R9 pol (50)
IP H LG SGLY FNPEW P H N DIIN C
Consensus (118)
176 190 200 210 220 236
(176)
P---SRNWKYLTPAKFWPKSISYFPVQVGVKPKYPDNVMQHESIVGKYLTRLY
DHBV pol(176)
QQFVGPLTVNEKRRLKLIMPARFYPTHTKYLPLDKGIKPYYPDQVVNHYFKTRHYLHTLW
HBV adw R9 pol (95)
R K I PAKFWP Y PL GIKP YPDNVMNH YL LW
Consensus (176)
235 240 250 260 270 280
(235)
YEAGILYKRISKHLVTFKGQPYNWEQQHLVNQHHIYDGATSSKINGR-QTDRRRRNTVK
DHBV pol(225)
WKAGILYKRETTRSASFCGSPYSWEQELQHGRLVIETSQRHGDESFCSQPSGILSRSSV
HBV adw R9 pol(154)
W AGILYKR S SF G PY WEQ I Q S
Consensus (235)
293 300 310 320 330 340
(293)
KPTCRKDDPKRDFDMVRQVSNTRSRVRPCANNGGDKHPPESGSLACWGGKES--RIIKS
DHBV pol(282)
VGPCIRSQLKQSRLGLQPHQGPLASSQPGRS-GSIRARVHPSTRRCFGVEPSGSGHIDH
HBV adw R9 pol(212) C K K L A P G K S CFG S I Consensus (293) 351 360 370 380 390 400 411 (351) SDSSRDSSAPVDSRGRPKSTRSFSPLSRRKTTGNHHHSSVFP-SSVEATTRG---RST DHBV pol(338)
HSVNNSSSCLHQSAVRKAAYSHLSTSKRQSSSGHAVEFHCLPPSSAGSQSQGSVFSCWWL
HBV adw R9 pol(269) SS S R A S R SSG P SS A S G Consensus (351) 410 420 430 440 450 460 (410) TPGKSVSPRDSSAIPVRTSGASDKNSPLEEENVWYLRGNTSWPNRITGKLFLVDKNSRN DHBV pol(391)
LQFRNSKPCSEYCLSHLVNLREDWG-PCDEYGEHHIR-IPRTPARVTGGVFLVDKNPHN
HBV adw R9 pol(328) K P I D P DE HIR P RITG LFLVDKN N Consensus (410) 1 10 20 30 40 50 61 (1) MQKLTRNHWIGLGDCFGGITTVYCGEKLKLLTIFLVCVLGCQLLRNIEVEMPRPLKQSLD DHBV pol (1)
-MPLSYQHFR---KLLLLDDGTEAGPLEEELP
HBV adw R9 pol (1) LS NHF LL D PL L Consensus (1) 59 70 80 90 100 119 (59) LDQSRWLREAEKQLRVLENLVDSNLEEEKLKPQLSMGEDVQSPGKGEPLHPNVRAPLSHV DHBV pol (59) LPR---LADADLNRRVAEDLNLGNL N---HBV adw R9 pol (27) L L DAD N RV E L NL Consensus (59) 118 130 140 150 160 (118)
VVRAATIDLPRLGNKLPARHHLGKLSGLYQMKGCTFNPEWKVPDISDTHFNLDVVNEC
DHBV pol(118)
---VSIPWTHKVGNFTGLYSSTVPIFNPEWQTPSFPKIHLQEDIINRC
HBV adw R9 pol (50)
IP H LG SGLY FNPEW P H N DIIN C
Consensus (118)
176 190 200 210 220 236
(176)
P---SRNWKYLTPAKFWPKSISYFPVQVGVKPKYPDNVMQHESIVGKYLTRLY
DHBV pol(176)
QQFVGPLTVNEKRRLKLIMPARFYPTHTKYLPLDKGIKPYYPDQVVNHYFKTRHYLHTLW
HBV adw R9 pol (95)
R K I PAKFWP Y PL GIKP YPDNVMNH YL LW
Consensus (176)
235 240 250 260 270 280
(235)
YEAGILYKRISKHLVTFKGQPYNWEQQHLVNQHHIYDGATSSKINGR-QTDRRRRNTVK
DHBV pol(225)
WKAGILYKRETTRSASFCGSPYSWEQELQHGRLVIETSQRHGDESFCSQPSGILSRSSV
HBV adw R9 pol(154)
W AGILYKR S SF G PY WEQ I Q S
Consensus (235)
293 300 310 320 330 340
(293)
KPTCRKDDPKRDFDMVRQVSNTRSRVRPCANNGGDKHPPESGSLACWGGKES--RIIKS
DHBV pol(282)
VGPCIRSQLKQSRLGLQPHQGPLASSQPGRS-GSIRARVHPSTRRCFGVEPSGSGHIDH
HBV adw R9 pol(212) C K K L A P G K S CFG S I Consensus (293) 351 360 370 380 390 400 411 (351) SDSSRDSSAPVDSRGRPKSTRSFSPLSRRKTTGNHHHSSVFP-SSVEATTRG---RST DHBV pol(338)
HSVNNSSSCLHQSAVRKAAYSHLSTSKRQSSSGHAVEFHCLPPSSAGSQSQGSVFSCWWL
HBV adw R9 pol(269) SS S R A S R SSG P SS A S G Consensus (351) 410 420 430 440 450 460 (410) TPGKSVSPRDSSAIPVRTSGASDKNSPLEEENVWYLRGNTSWPNRITGKLFLVDKNSRN DHBV pol(391)
LQFRNSKPCSEYCLSHLVNLREDWG-PCDEYGEHHIR-IPRTPARVTGGVFLVDKNPHN
HBV adw R9 pol(328) K P I D P DE HIR P RITG LFLVDKN N Consensus (410) DTN DTC DS DRN
24
(continued)
468 480 490 500 510 528
(468)
NTEEARLVVDFSQFSKGKNAMRFPRYWSPNLSTLRRILPVGMPRISLDLSQAFYHLPLNP
DHBV pol(449)
NTAESRLVVDFSQFSRGITRVSWPKFAVPNLQSLTNLLSSNLSWLSLDVSAAFYHIPLHP
HBV adw R9 pol(384)
NT EARLVVDFSQFSKG M FPKF PNL SL IL L ISLDLS AFYHIPL P
Consensus (468) 527 540 550 560 570 (527) PASSSRLAV SDG---DHBV pol(508) PAAMPHLLVGSSGLSRYVARLSSNSRNNNNQYGTMQNLHDSCSRQLYVSLMLLYKTFGW HBV adw R9 pol(443) PAA L V Consensus (527) 585 590 600 610 620 630 (585) ---QR---VYYFRKAPMGVGLSPFLLHLFTTALGSEISRRFN-VWTFTYMDDFLLCH DHBV pol(520)
WKLHLYSHPIVLGFRKIPMGVGLSPFLLAQFTSAICSVVRRAFPHCLAFSYMDDVVLGA
HBV adw R9 pol(501)
V FRK PMGVGLSPFLL FTSAI S I R F FSYMDD LL
Consensus (585)
643 650 660 670 680 690 703
(643)
HPNARHLNAISHAVCSFLQELGIRINFDKTTPSPVNEIRFLGYQIDENFMKIEESRWKE
DHBV pol(569)
AKSVQHRESLYTAVTNFLLSLGIHLNPNK-TKRWGYSLNFMGYIIGSWGTLPQDHIVQK
HBV adw R9 pol(559)
H AI AV FL LGI IN K T I FLGY I D
Consensus (643)
702 710 720 730 740 750 762
(702)
LRTVIKKIKVGEWYDWKCIQRFVGHLNFVLPFTKGNIEMLKPMYAAITNQVNFSFSSSYRT
DHBV pol(628)
IKHCFRKLPVNRPIDWKVCQRIVGLLGFAAPFTQCGYPALMPLYACIQAKQAFTFSPTYKA
HBV adw R9 pol(617)
IK KKI V DWK QR VG L F PFT L PLYA I FSFS SYK
Consensus (702)
760 770 780 790 800 810 820
(760)
YRTLLYKLTMGVCKLRIKPKSSVPLPRVATDATPTHGAISHITGGSAVFAFSKVRDIHV
DHBV pol(686)
YKAFLSKQYMNLYPVARQRPG---LCQVFADATPTGWGLA-IGHQRMRGTFVAPLPIHT
HBV adw R9 pol(675)
YK L K M L L L V DATPT AIA I F IH
Consensus (760)
819 830 840 850 860
(819)
QELLMSCLAKIMIKPRCLLSDSTFVCHKRYQTLPWHFAMLAKQLLKPIQLYFVPSKY
DHBV pol(745)
AELLAACFARSRSGAKLIGTDNSVVLSRKYTSFPWLLGCTANWILRGTSFVYVPSAL
HBV adw R9 pol(730)
ELL AC AK K I SD S V KKY S PW A A ILK FVPS
Consensus (819)
875 880 890 900 910 920 935
(875)
YNPADGPSR---HKPPDWTAFPYTPLSKAIY
IPHRLCGT---DHBV pol(801) LNPADDPSRGRLGLSRPLLRLPFQPTTGRTSLY AVSPSVPSHLPVRVHFASPLHVAWRPP-HBV adw R9 pol(786) NPAD PSR KP F T AIY S Consensus (875) DRC DHN DH1C DH2C 468 480 490 500 510 528 (468)
NTEEARLVVDFSQFSKGKNAMRFPRYWSPNLSTLRRILPVGMPRISLDLSQAFYHLPLNP
DHBV pol(449)
NTAESRLVVDFSQFSRGITRVSWPKFAVPNLQSLTNLLSSNLSWLSLDVSAAFYHIPLHP
HBV adw R9 pol(384)
NT EARLVVDFSQFSKG M FPKF PNL SL IL L ISLDLS AFYHIPL P
Consensus (468) 527 540 550 560 570 (527) PASSSRLAV SDG---DHBV pol(508) PAAMPHLLVGSSGLSRYVARLSSNSRNNNNQYGTMQNLHDSCSRQLYVSLMLLYKTFGW HBV adw R9 pol(443) PAA L V Consensus (527) 585 590 600 610 620 630 (585) ---QR---VYYFRKAPMGVGLSPFLLHLFTTALGSEISRRFN-VWTFTYMDDFLLCH DHBV pol(520)
WKLHLYSHPIVLGFRKIPMGVGLSPFLLAQFTSAICSVVRRAFPHCLAFSYMDDVVLGA
HBV adw R9 pol(501)
V FRK PMGVGLSPFLL FTSAI S I R F FSYMDD LL
Consensus (585)
643 650 660 670 680 690 703
(643)
HPNARHLNAISHAVCSFLQELGIRINFDKTTPSPVNEIRFLGYQIDENFMKIEESRWKE
DHBV pol(569)
AKSVQHRESLYTAVTNFLLSLGIHLNPNK-TKRWGYSLNFMGYIIGSWGTLPQDHIVQK
HBV adw R9 pol(559)
H AI AV FL LGI IN K T I FLGY I D
Consensus (643)
702 710 720 730 740 750 762
(702)
LRTVIKKIKVGEWYDWKCIQRFVGHLNFVLPFTKGNIEMLKPMYAAITNQVNFSFSSSYRT
DHBV pol(628)
IKHCFRKLPVNRPIDWKVCQRIVGLLGFAAPFTQCGYPALMPLYACIQAKQAFTFSPTYKA
HBV adw R9 pol(617)
IK KKI V DWK QR VG L F PFT L PLYA I FSFS SYK
Consensus (702)
760 770 780 790 800 810 820
(760)
YRTLLYKLTMGVCKLRIKPKSSVPLPRVATDATPTHGAISHITGGSAVFAFSKVRDIHV
DHBV pol(686)
YKAFLSKQYMNLYPVARQRPG---LCQVFADATPTGWGLA-IGHQRMRGTFVAPLPIHT
HBV adw R9 pol(675)
YK L K M L L L V DATPT AIA I F IH
Consensus (760)
819 830 840 850 860
(819)
QELLMSCLAKIMIKPRCLLSDSTFVCHKRYQTLPWHFAMLAKQLLKPIQLYFVPSKY
DHBV pol(745)
AELLAACFARSRSGAKLIGTDNSVVLSRKYTSFPWLLGCTANWILRGTSFVYVPSAL
HBV adw R9 pol(730)
ELL AC AK K I SD S V KKY S PW A A ILK FVPS
Consensus (819)
875 880 890 900 910 920 935
(875)
YNPADGPSR---HKPPDWTAFPYTPLSKAIY
IPHRLCGT---DHBV pol(801) LNPADDPSRGRLGLSRPLLRLPFQPTTGRTSLY AVSPSVPSHLPVRVHFASPLHVAWRPP-HBV adw R9 pol(786) NPAD PSR KP F T AIY S Consensus (875) 468 480 490 500 510 528 (468)
NTEEARLVVDFSQFSKGKNAMRFPRYWSPNLSTLRRILPVGMPRISLDLSQAFYHLPLNP
DHBV pol(449)
NTAESRLVVDFSQFSRGITRVSWPKFAVPNLQSLTNLLSSNLSWLSLDVSAAFYHIPLHP
HBV adw R9 pol(384)
NT EARLVVDFSQFSKG M FPKF PNL SL IL L ISLDLS AFYHIPL P
Consensus (468) 527 540 550 560 570 (527) PASSSRLAV SDG---DHBV pol(508) PAAMPHLLVGSSGLSRYVARLSSNSRNNNNQYGTMQNLHDSCSRQLYVSLMLLYKTFGW HBV adw R9 pol(443) PAA L V Consensus (527) 585 590 600 610 620 630 (585) ---QR---VYYFRKAPMGVGLSPFLLHLFTTALGSEISRRFN-VWTFTYMDDFLLCH DHBV pol(520)
WKLHLYSHPIVLGFRKIPMGVGLSPFLLAQFTSAICSVVRRAFPHCLAFSYMDDVVLGA
HBV adw R9 pol(501)
V FRK PMGVGLSPFLL FTSAI S I R F FSYMDD LL
Consensus (585)
643 650 660 670 680 690 703
(643)
HPNARHLNAISHAVCSFLQELGIRINFDKTTPSPVNEIRFLGYQIDENFMKIEESRWKE
DHBV pol(569)
AKSVQHRESLYTAVTNFLLSLGIHLNPNK-TKRWGYSLNFMGYIIGSWGTLPQDHIVQK
HBV adw R9 pol(559)
H AI AV FL LGI IN K T I FLGY I D
Consensus (643)
702 710 720 730 740 750 762
(702)
LRTVIKKIKVGEWYDWKCIQRFVGHLNFVLPFTKGNIEMLKPMYAAITNQVNFSFSSSYRT
DHBV pol(628)
IKHCFRKLPVNRPIDWKVCQRIVGLLGFAAPFTQCGYPALMPLYACIQAKQAFTFSPTYKA
HBV adw R9 pol(617)
IK KKI V DWK QR VG L F PFT L PLYA I FSFS SYK
Consensus (702)
760 770 780 790 800 810 820
(760)
YRTLLYKLTMGVCKLRIKPKSSVPLPRVATDATPTHGAISHITGGSAVFAFSKVRDIHV
DHBV pol(686)
YKAFLSKQYMNLYPVARQRPG---LCQVFADATPTGWGLA-IGHQRMRGTFVAPLPIHT
HBV adw R9 pol(675)
YK L K M L L L V DATPT AIA I F IH
Consensus (760)
819 830 840 850 860
(819)
QELLMSCLAKIMIKPRCLLSDSTFVCHKRYQTLPWHFAMLAKQLLKPIQLYFVPSKY
DHBV pol(745)
AELLAACFARSRSGAKLIGTDNSVVLSRKYTSFPWLLGCTANWILRGTSFVYVPSAL
HBV adw R9 pol(730)
ELL AC AK K I SD S V KKY S PW A A ILK FVPS
Consensus (819)
875 880 890 900 910 920 935
(875)
YNPADGPSR---HKPPDWTAFPYTPLSKAIY
IPHRLCGT---DHBV pol(801) LNPADDPSRGRLGLSRPLLRLPFQPTTGRTSLY AVSPSVPSHLPVRVHFASPLHVAWRPP-HBV adw R9 pol(786) NPAD PSR KP F T AIY S Consensus (875) DRC DHN DH1C DH2C
25
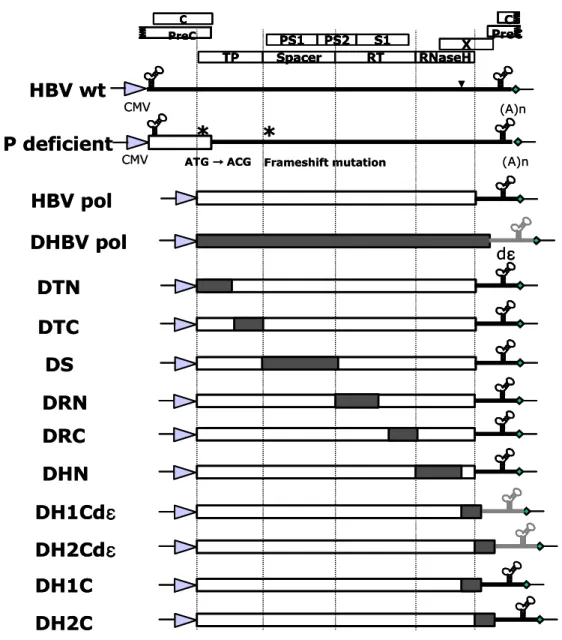
B. HBV and DHBV chimeric DNA polymerases constructions
Chimeric DNA polymerases were constructed to map the essential motif of DNA polymerase that support encapsidation or replication (Fig. 2). Since HBV wt construct has longer than genome length (Kim et al., 2004), this construct could support the pgRNA and mRNA expressions and supply all proteins required for encapsidation and replication. Polymerase deficient mutant, P deficient, could express pgRNA and core, surface, and X protein except polymerase (Kim et al., 2004). This P deficient mutant construct was used to co-transfect with various DNA polymerases including chimeric DNA polymerases to map the essential motifs of DNA polymerase. For the constructions of chimeric DNA polymerases of HBV and DHBV, amino acid sequence of DNA polymerase was aligned and then four domains of hepadnavirus polymerase were subdivided into N- and C-terminus (Fig. 1). Chimeric DNA polymerases were generated by fusion PCR techniques. Then each construct is named according the exchanged region of DHBV polymerase. Spacer region was not divided calling pDS construct. Since 3'-end sequences of HBV and DHBV may influence the polymerase expressions, DH1Cdε and DH2Cdε constructs that contain 3'-end sequences of DHBV were included.
26
Fig. 2. Schematic diagram of chimeric DNA polymerases construct with substituted DHBV polymerase sequence. Longer than genome length of HBV adw
R9 DNA was inserted into downstream of cytomegalovirus immediate early (CMV IE) promoter. For P deficient mutant construct, mutated sites are marked with asterisk. Open boxes and hatched boxes indicate HBV and DHBV sequences, respectively. C PS1 PS2 S1 X TP Spacer RT RNaseH C PreC PreC CMV (A)n
*
*
ATG →→→ ACG→ Frameshift mutation P deficient HBV wt CMV (A)n HBV pol DHBV pol dεεεε DTN DTC DS DRN DRC DH1Cdεεεε DH2Cdεεεε DH1C DH2C DHN C PS1 PS2 S1 X TP Spacer RT RNaseH C PreC PreC C PS1 PS2 S1 X TP Spacer RT RNaseH C PreC PreC CMV (A)n
*
*
ATG →→→ ACG→ Frameshift mutation P deficient HBV wt CMV (A)n HBV pol DHBV pol dεεεε DTN DTC DS DRN DRC DH1Cdεεεε DH2Cdεεεε DH1C DH2C DHN
27
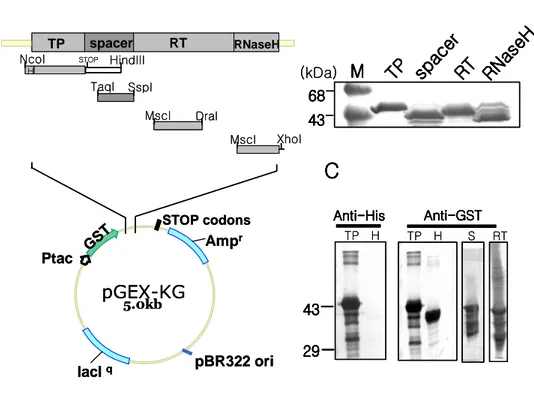
C. Generation of polyclonal antibody specific for each domain of HBV DNA polymerase expressed from E. coli
Although all previous attempts to generate full-length active polymerase had been failed, the expressions of separated domains were possible (Beck and Nassal, 2001). On the basis of this point, separated TP, spacer, RT and RNase H domains were expressed in MC1061 strain of E. coli. As shown in Fig. 3A, E. coli MC1061 cells were transformed with fusion constructs of each domain, induced, and produced large amounts of the TP, spacer, RT and RNase H fusion proteins with approximate molecular weight of 49, 43.5, 46, and 42 KDa, respectively (Fig. 3B). Since lager proportions of fusion proteins were present as insoluble inclusion bodies, the fusion proteins were purified from the sliced SDS-PAGE gel. Western blot analysis with specific antibodies against to GST, each fusion proteins (Fig. 3C) were expressed, as expected, but small sized protein bands were shown. Such small proteins are supposed to be degraded proteins during gel purification. Each rabbit per fusion protein was subcutaneously and intradermally immunized with gel-purified fusion proteins for 6 months with several boosts. Finally, the serum of each immunized animal exhibited a strong reactivity against fusion protein antigens at 1:32,000 dilutions. All polyclonal rabbit antiserum worked well in Western blotting, but only TP specific antisera can recognize DNA polymerase in transfected cells, not other polyclonal rabbit antiserum. This TP specific antiserum will be used in this study as shown below.
28
Fig. 3. The bacterial expression and the purification of GST-fused HBV polymerase domains. (A) Schematic diagram of plasmid constructs for bacterial
expression. The domain of the polymerase that is expressed in E. coli is shown in relative positions on polymerase open reading frame. TP, spacer, RT and RNaseH domains are indicated. TP has His tag (hatched box). (B) The expressions of HBV DNA polymerase domains in E. coli MC1061. Purified fractions were resolved by SDS-PAGE and stained with Coomassie brilliant blue. (C) Western blot of fusion protein. The first mouse anti-GST or anti-His antibodies, secondary AP-conjugated anti-mouse antibody, and PNPP substrate were used to visualize proteins by Western blot analysis. pGEX-KG 5.0kb Ampr lacIq Ptac pBR322 ori GST STOP codons RT RNaseH spacer TP
NcoI STOP HindIII TaqI SspI MscI DraI MscI XhoI H
A
A
A
A
B
B
B
B
C
C
C
C
TP H TP H S RT Anti AntiAntiAnti----HisHisHisHis Anti-AntiAntiAnti---GSTGSTGSTGST
43 43 43 43 29 29 29 29 43 43 43 43 68 6868 68 TP TPTP TP spac er spac er spac er spac er RT RTRT RT RNas eH RNas eH RNas eH RNas eH M MM M (kDa) pGEX-KG 5.0kb Ampr lacIq Ptac pBR322 ori GST STOP codons RT RNaseH spacer TP
NcoI STOP HindIII TaqI SspI MscI DraI MscI XhoI H
A
A
A
A
B
B
B
B
C
C
C
C
TP H TP H S RT Anti AntiAntiAnti----HisHisHisHis Anti-AntiAntiAnti---GSTGSTGSTGST
43 43 43 43 29 29 29 29 43 43 43 43 68 6868 68 TP TPTP TP spac er spac er spac er spac er RT RTRT RT RNas eH RNas eH RNas eH RNas eH M MM M (kDa)
29
D. Chimeric DNA polymerases constructs cannot support the encapsidation and the replication of HBV
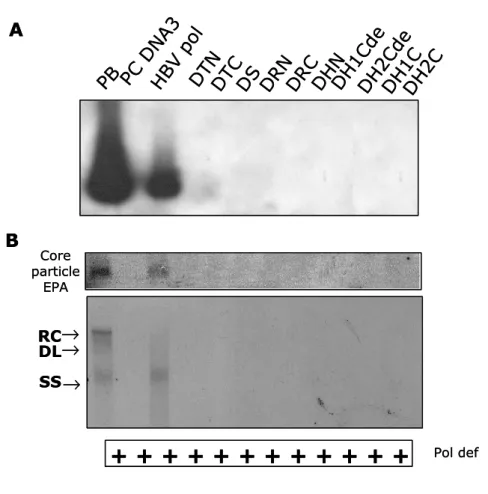
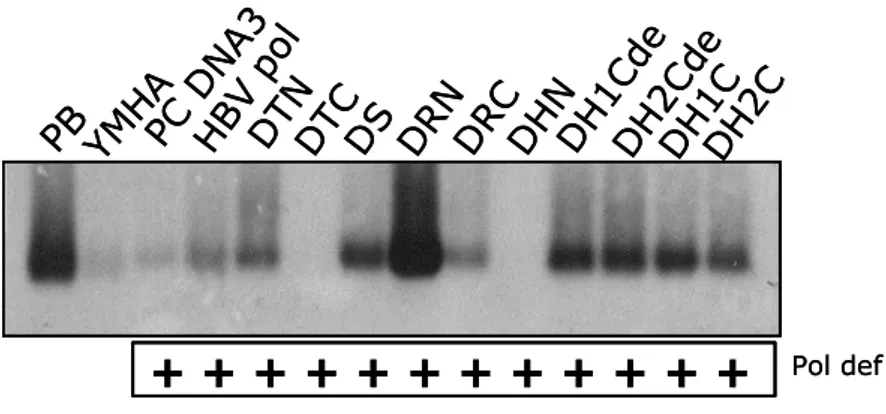
To identify the domains of DNA polymerase that is essential for encapsidation or replication, encapsidation assay (Fig. 4A) and endogenous polymerase assay (Fig. 4B, upper and lower panel) were performed. HuH7 cells were co-transfected with chimeric DNA polymerase constructs and P deficient mutant construct that provides pgRNA and all of HBV proteins except DNA polymerase. HBV wt construct, pPB, which provides polymerase in cis and provides all of the requirements to support HBV replication, is used as positive control. pPB transfected cells and wt HBV polymerase and P deficient mutant co-transfected cells exhibited endogenous DNA polymerase activity and encapsidated HBV nucleic acids including pgRNA and HBV DNA. After EPA, core particles were subjected on a native agarose gel electrophoresis (Fig. 4B. upper panel) or nucleic acids in core particles were extracted and separated by 1% agarose gel electrophoresis to detect single-, double-stranded, and partially double-stranded relaxed circular forms of HBV DNA synthesis, if any (Fig. 4B. lower panel). However all chimeric DNA polymerase cannot trans-complement P deficient mutant in which pgRNA encapsidation and replication rescue of P deficient mutant were not observed.
30
Fig. 4. Chimeric DNA polymerase constructs cannot support HBV replications.
(A) Encapsidation assay to identify the HBV nucleic acids in situ from disrupted core particles. P deficient mutant was co-transfected with chimeric DNA polymerases construct. Fixed nucleic acids were hybridized with random-primed [32P] labeled HBV specific probes and subjected to autoradiography. (B) Endogenous polymerase assay (EPA). Isolated core particles were supplemented with 10 µCi of α- [32P]-dATP (3,000 Ci/mmol) and unlabeled dNTP mix in EPA reaction buffer, electrophoresed on a 1% native agarose gel, and subjected to autoradiography (upper panel). [32 P]-labeled DNA after EPA was extracted, separated, and subjected to autoradiography (lower panel). Single-, double-stranded, and partially double-stranded relaxed circular forms of HBV DNA are marked as SS, DS, and RC, respectively. All chimeric DNA polymerases were designated according to the substituted portion of DHBV DNA polymerase. For example, when N-terminal region of terminal protein of DHBV DNA polymerase was substituted into HBV DNA polymerase, chimeric DNA polymerase was designated as DTN. Abbreviation: T, terminal protein; S, spacer; R, reverse transcriptase; H, RNaseH.
31
Fig. 4. Chimeric DNA polymerase constructs cannot support HBV replications. PB PC DN A3 HB V p ol DT N DT C DS DR N DR C DH N DH 1C de DH 2C de DH 1C DH 2C RC DL SS
+ + + +
+
+
+ +
+
+
+ +
Pol def → → → A B Core particle EPA PB PC DN A3 HB V p ol DT N DT C DS DR N DR C DH N DH 1C de DH 2C de DH 1C DH 2C RC DL SS+ + + +
+
+
+ +
+
+
+ +
Pol def → → → A B Core particle EPA32
E. Chimeric DNA polymerase RNA expression
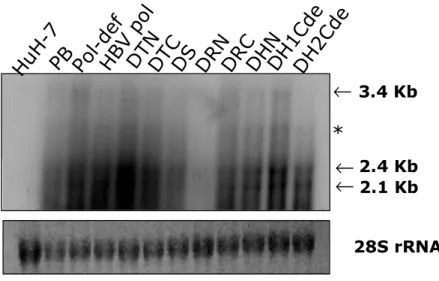
To investigate the reason why chimeric DNA polymerase cannot support HBV encapsidation and replication, the chimeric RNAs and chimeric polymerases expressions were analyzed. pPB, wt HBV DNA polymerase or chimeric DNA polymerase construct were transfected into the HuH7 cells respectively. Total RNA was analyzed by Northern blot analysis. Upon transfection, pgRNA was expressed by the action of the CMV promoter and subgenomic RNAs were synthesized under the control of their authentic promoters. The 3.5 Kbp pgRNA, and 2.4 and 2.1 Kbp mRNAs of surface proteins were detected from the pPB and P deficient mutant transfected cells (Fig. 5). A bit smaller RNA than pgRNA would be expected for chimeric polymerases but there were no significant size differences between pgRNA and chimeric DNA polymerase RNAs. It is possible that the size differences between pgRNA and chimeric DNA polymerase RNAs may not be as apparent in Northern blot analysis. There was another RNA band marked as asterisk between pgRNA and subgenomic RNA that may be a spliced RNA (Fig 5). In DRN construct transfected cells chimeric RNA and subgenomic RNA were very low. However, using the X gene specific probe, chimeric RNA and subgenomic RNA in DRN transfected cells were detected (data not shown) and showed similar RNA level from other constructs transfected cells indicating that this very low level might be due to the probe used to detect HBV sequences in this experiment, since the probe contain most of HBV RT sequences.
33
Fig. 5. HBV RNA expressions from chimeric DNA polymerase transfected HuH7 cells. Northern blot analysis was performed to detect HBV subgenomic mRNA and
chimeric HBV DNA polymerase mRNA. Total RNA was extracted and separated by 1% formaldehyde gel electrophoresis, transferred to nylon membranes, hybridized with random-primed 32P labeled HBV specific probes, and subjected to autoradiography. Chimeric HBV RNA (3.4 Kbp) and 2.1 and 2.4 Kbp mRNAs for surface protein(s) are indicated by arrows. Spliced RNAs from chimeric HBV RNA are indicated by asterisk.
H
u
H
-7
PB Po
l-d
ef
H
B
V
p
o
l
D
T
N
D
T
C
D
S
D
R
N
D
R
C
D
H
N
D
H
1
C
d
e
D
H
2
C
d
e
3.4 Kb 2.4 Kb 2.1 Kb 28S rRNA←
←
←
*
34
F. HBV DNA polymerase expression
By immunofluorescence assay, intracellular distribution of the HBV DNA polymerase was analyzed from various polymerase constructs transfected HuH7 cells with TP-specific antibody that had been prepared in this study (Fig. 2) and endoplasmic reticulum (ER) specific antibodies, Golgi specific antibodies, or peroxisome specific antibodies. The polymerase was readily detectable in the cytoplasm of transfected HuH7cells as bright red color. The merge of the images of polymerase and ER, Golgi, or peroxisome revealed that HBV polymerase are not co-localized with these organelles and distributions of polymerase in cytoplasm differ from that of surface antigen (Fig. 6A). These results suggest that similar to DHBV, major of HBV polymerase may have another function such as pathological involvement besides the functions in virus replication and encapsidation. By immunofluorecence assay with TP specific polyclonal antiserum, HBV DNA polymerase detection from HBV wt transfected cells was possible but the expression signal was weaker than HBV polymerase only expressed cells. This result was consistent with the recent report that non-recombinant HBV polymerase from HBV replicating cells were very difficult to detect, since from which core proteins are expressed and polymerase are encapsidated in core particles (Yao et al., 2000). During virus life cycle, viral particles containing viral proteins and nucleic acids move from the site of replication to that of virus assembly so that they have to either hijack the cytoplasmic vesicle traffic or to interact directly with the cytoskeletal
35
transport machinery to be transported to plasma membrane (Dohner and Sodeik, 2005). Double immuno-staining was performed to understand the relationship between HBV DNA polymerase and microtubule. HBV polymerase was not co-localized with microtubule (Fig. 5Bb). This results suggest that most of cytoplasmic HBV DNA polymerase might play another role besides the participation in encapsidation since the encapsidated core particle trafficking must utilize the host cytoskeletal system even though direct interaction have not been identified yet in HBV. By immunofluorescence assay, HBV DNA polymerase and core protein did not seem to interact directly since HBV DNA polymerase and other HBV proteins localized separately (data not shown). When DNA polymerase and surface protein were double immuno-stained with TP specific antibody and surface antigen specific antibody, respectively, these proteins seemed to be co-localized (Fig. 5Ba). This result shows that the PS (polymerase-surface) fusion protein also is expressed in HBV replicating cells. This aspect will be further explored in part II. However, chimeric DNA polymerases could not detected by immunoflorescence assay.