저작자표시-비영리-변경금지 2.0 대한민국 이용자는 아래의 조건을 따르는 경우에 한하여 자유롭게 l 이 저작물을 복제, 배포, 전송, 전시, 공연 및 방송할 수 있습니다. 다음과 같은 조건을 따라야 합니다: l 귀하는, 이 저작물의 재이용이나 배포의 경우, 이 저작물에 적용된 이용허락조건 을 명확하게 나타내어야 합니다. l 저작권자로부터 별도의 허가를 받으면 이러한 조건들은 적용되지 않습니다. 저작권법에 따른 이용자의 권리는 위의 내용에 의하여 영향을 받지 않습니다. 이것은 이용허락규약(Legal Code)을 이해하기 쉽게 요약한 것입니다. Disclaimer 저작자표시. 귀하는 원저작자를 표시하여야 합니다. 비영리. 귀하는 이 저작물을 영리 목적으로 이용할 수 없습니다. 변경금지. 귀하는 이 저작물을 개작, 변형 또는 가공할 수 없습니다.
Master's Thesis in the Department of
Biomedical Sciences
Circulating Exosomal MicroRNA-1307-5p as a
Predictive marker for Metastasis in
hepatocellular carcinoma
Ajou University Graduate School
Major in Cancer Biology
Circulating Exosomal MicroRNA-1307-5p as a
Predictive marker for Metastasis in
hepatocellular carcinoma
Jae Youn Cheong, Advisor.
I submit this thesis as the Master's thesis
in the Department of Biomedical Sciences.
August, 2020
Ajou University Graduate School
Major in Cancer Biology
i
ABSTRACT
Circulating Exosomal MicroRNA-1307-5p as a Predictive marker
for Metastasis in hepatocellular carcinoma
Background/purpose : Exosomal microRNAs (exo-miRs) have been reported to play an important role in cancer metastasis. This study aimed to identify pro-metastatic circulating exo-miRs in hepatocellular carcinoma (HCC).
Methods : NGS-based plasma exo-miR profile was analyzed in 14 patients with HCC, consisted of 8 patients without metastasis and 6 patients with metastasis within 1 year of follow up. Integrative analyses were performed to select candidate miRs using two different public expression datasets-GSE67140 and The Cancer Genomic Atlas Liver Hepatocellular Carcinoma (TCGA_LIHC). Validation was performed in stored blood sample of 150 HCC patients at the time of diagnosis. Downstream signaling pathway of selected miR was predicted by Targetscan and Ingenuity pathway analysis (IPA).
Results : A total of 61 miRs were significantly overexpressed in patients with metastasis. Integrative analyses with public datasets identified 3 of 61 miRs, miR-106b-5p, miR-1307-5p, and miR-340-miR-1307-5p, which commonly overexpressed both in metastasis and vascular invasion group and showed prognostic implication. In validation study, circulating
ii
miR-1307-5p was overexpressed in metastasis group (P=0.04). It was also significantly overexpressed in patients with vascular invasion and tumor recurrence. The expression of circulating exo-miR-1307-5p was correlated with the progression of tumor stage (P<0.0001). In a comprehensive bioinformatics analysis, the downstream pathway of miR-1307-5p, which promoting epithelial mesenchymal transmission (EMT), was proposed as a down-regulation of SEC14L2 and ENG.
Conclusion : This study suggested circulating exo-miR-1307-5p as a metastasis driver molecule and a predictive marker for the occurrence of metastasis in HCC patients. SEC14L2 and ENG were predicted as target tumor suppressor gene of miR-1307, which promoting EMT.
key words : Hepatocellular carcinoma, metastasis, exosome, microRNA, bioinformatics analysis
iii
TABLES OF CONTENTS
ABSTRACT ∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙ i TABLE OF CONTENTS ∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙ iii LIST OF FIGURES ∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙ v LIST OF TABLES ∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙ iii I. INTRODUCTION ∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙ 1 II. MATERIALS AND METHODS ∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙ 3
1. Patients and samples collection ∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙ 3
2. GEO database and TCGA_LIHC database analysis ∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙ 6
3. Blood exosome Isolation and Total exosomal RNA Extraction ∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙ 6
4. Small RNA sequencing ∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙ 6
5. Transmission electron microscopy ∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙ 7
6. Nanoparticle tracking analysis ∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙ 7
7. Western Blot ∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙ 8
8. Quantitative Real-time Polymerase chain reaction. (RT-qPCR) ∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙ 8
9. Target prediction of miRNA. ∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙ 11
10. Ingenuity pathway analysis ∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙ 11
11. Statistical analysis ∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙ 11 III. RESULTS ∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙ 12
iv
in metastasis group ∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙ 12
2. Integrative analysis with public gene expression datasets to select potential candidates of pro-metastatic miR ∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙ 15
3. Validating clinical implication of the candidate exo-miRs in the validation cohort ∙∙∙∙∙∙ 20
4. Predicting target genes of miR-1307-5p in HCC patients using bioinformatics analysis 23 IV. DISCUSSION ∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙ 29 V. REFERENCES ∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙ 33
v
LIST OF FIGURES
Figure 1. Pipeline of the systematic integrative analyses performed in the present study to identify metastasis driver circulating exosomal microRNAs ∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙ 13 Figure 2. Confirmation of isolated circulating exosomes and identification of overexpressed exo-miRs in metastasis group ∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙ 14 Figure 3. Integrative analyses with two different public RNA sequencing datasets ∙∙∙∙∙∙ 17 Figure 4. Kaplan-Meier plot of overall survival according to expression of microRNA-193, 202, 33b, 455, 542, and microRNA-574 in TCGA_LIHC ∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙ 19 Figure 5. Validation of clinical implication of the candidate exosomal microRNAs in the validation cohort ∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙ 21 Figure 6. Predicting target genes of microRNA-1307-5p in hepatocellular carcinoma using bioinformatics analysis ∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙ 25 Figure 7. Expression of the 9 target gene candidates of microRNA-1307-5p in TCGA_LIHC ∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙27 Figure 8. Pathway analysis with a functional of the EMT using the IPA software on
vi
LIST OF TABLES
Table 1. Baseline characteristics of the plasma-HCC cohort and Serum-HCC cohort ∙∙∙∙∙∙∙∙∙∙∙∙∙∙ 5
Table 2. miRNA sequence used for qRT-PCR ∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙∙ 10
1
I. INTRODUCTION
Hepatocellular carcinoma (HCC) is the sixth most common malignancy and the third leading cause of cancer-related mortality worldwide. (1) During the last several decades, the prognosis of HCC has been much improved as a result of advances in diagnostic and therapeutic approaches of HCC. (2) However, the prognosis of HCC patients in the advanced stage remains very poor with median survival of 4-6 months. (3) In HCC patients, presence of metastasis is a key determinant of treatment strategy, as locoregional therapies are no longer effective to control extrahepatic metastasis. (4) Therefore, determining metastatic status during the initial staging process is essential for planning an appropriate treatment strategy, which is directly related to survival. Majority of extrahepatic metastases are usually detected by conventional imaging modalities such as computed tomography and bone scintigraphy. However, it requires considerable effort and cost, and sometimes it cannot detect small lesions of metastasis. Detection of metastasis driver molecules prior to the diagnosis by conventional imaging technique will help classifying patients according to the stratified risk of metastasis, which would facilitate implementation of precision medicine.
Liquid biopsy is a technology for detection of tumor-derived genetic molecules in body fluid, such as blood, urine and saliva. (5-7) Liquid biopsy can target various classes of circulating tumor molecules including cell free DNA, circulating tumor cells, and tumor cell derived extra-cellular vesicles. (8) Exosomes are about 30–100 nm sized
2
extracellular vesicles, which carry genetic components of the parent cell to the recipient cell. (9, 10) Exosomal contents have been studied as major target of liquid biopsy. (11) Among the genetic materials of exosome, microRNAs (miRs) have been spotlighted because several publications suggest that the loading of specific miRs into exosome is actively selected procedure according to property of parent cell. (12, 13) Recently, exosomes have been reported as instigators of metastasis niche formation by transfer functional molecules activating diverse epithelial mesenchymal transition (EMT) promoting signaling pathway in recipient cells. (14, 15) In particular, exosomal miRs (exo-miRs) have been reported to play an important role in cancer metastasis. However, especially in HCC, exosome as a metastasis mediator is still in its infancy despite breakthrough in the field of liquid biopsy over the past few years.
In this study, we aimed to identify pro-metastatic circulating exo-miRs, which could be used as a metastasis predictor in HCC by comprehensive and systematic integrative analyses of plasma exo-miRs sequencing data and publicly available RNA sequencing (RNA-seq) datasets. We also aimed to propose a possible mechanism of pro-metastatic miRs, including EMT process.
3
II. MATERIAL & METHOD
1. Patients and sample collection
Blood samples were provided by Biobank of Ajou University Hospital, a member of the Korea Biobank Network. To identify candidates of circulating exo-miRs with pro-metastatic potential, medical records of HCC patients with available plasma sample at the time of diagnosis were reviewed and enrolled according to inclusion and exclusion criteria. The inclusion criteria were as following: 1. newly diagnosed HCC patients according to AASLD criteria, (16) 2. without extrahepatic metastasis at the time of diagnosis, 3. treated with local or systemic therapy according to tumor burden or location, (17) 4. Child-Pugh class A or B, and 5. presence of followed-up data with imaging study for evaluating tumor burden and metastasis status every 3 months more than 1 year. The patients, who were lost to follow-up before 1 year without metastasis event, were excluded. Among 52 patients who met the inclusion criteria, extrahepatic metastasis was occurred in 25 patients (metastasis group), while 27 patients were remained as metastasis-free during follow-up period (metastasis-free group). This cohort was named as Plasma-HCC cohort. Table 1 demonstrates baseline characteristics of the Plasma-HCC cohort.
Due to the limited number and lack of several clinical information in the Plasma-HCC cohort, Plasma-HCC patients with available pre-treatment serum sample at the time of
4
diagnosis were included to validate clinical implication of selected circulating exo-miRs. This validation cohort was named as Serum-HCC cohort. The Serum-HCC cohort consisted of 91 serum samples categorized as 73 patients with HCC and 18 normal healthy controls. A healthy control was defined as an individual without any medical history, who visited the Ajou Health Promotion Center for a regular health check-up. The vascular invasion status, metastasis status, and the tumor stage according to the modified Union for International Cancer Control (mUICC) staging system were obtained. The baseline characteristics of the Serum-HCC cohort was demonstrated in Table 1.
The study protocol was approved by the Institutional Review Board of the Ajou University Hospital, Suwon, South Korea (AJRIB-BMR-OBS-16-344). Anonymous blood samples and clinical data were provided by the Ajou Human Bio-Resource Bank. Informed consent was waived.
5
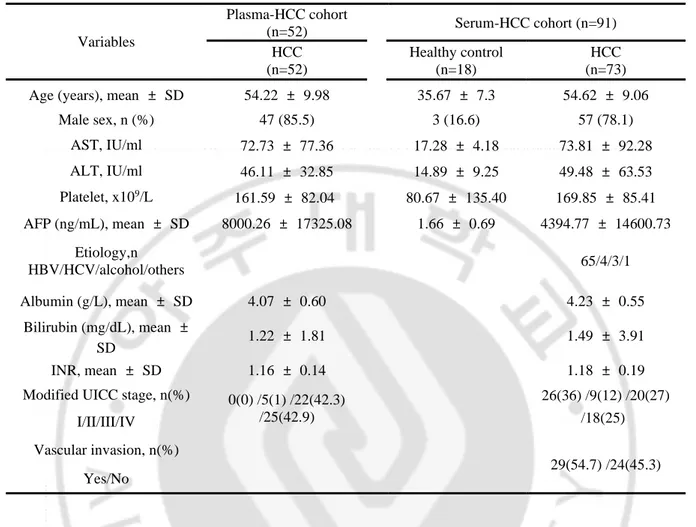
Table 1. Baseline characteristics of the Plasma-HCC cohort and Serum-HCC cohort
HCC, hepatocellular carcinoma; AST , astpartate transaminase; ALT, alanine transaminase; AFP, alpha-feto protein; HBV, hepatitis B virus; HCV, hepatitis C virus; INR, international normalized ratio; UICC, Union for International Cancer Control
Variables Plasma-HCC cohort (n=52) Serum-HCC cohort (n=91) HCC (n=52) Healthy control (n=18) HCC (n=73) Age (years), mean ± SD 54.22 ± 9.98 35.67 ± 7.3 54.62 ± 9.06
Male sex, n (%) 47 (85.5) 3 (16.6) 57 (78.1) AST, IU/ml 72.73 ± 77.36 17.28 ± 4.18 73.81 ± 92.28 ALT, IU/ml 46.11 ± 32.85 14.89 ± 9.25 49.48 ± 63.53 Platelet, x109/L 161.59 ± 82.04 80.67 ± 135.40 169.85 ± 85.41 AFP (ng/mL), mean ± SD 8000.26 ± 17325.08 1.66 ± 0.69 4394.77 ± 14600.73 Etiology,n HBV/HCV/alcohol/others 65/4/3/1 Albumin (g/L), mean ± SD 4.07 ± 0.60 4.23 ± 0.55 Bilirubin (mg/dL), mean ± SD 1.22 ± 1.81 1.49 ± 3.91 INR, mean ± SD 1.16 ± 0.14 1.18 ± 0.19
Modified UICC stage, n(%) 0(0) /5(1) /22(42.3) /25(42.9) 26(36) /9(12) /20(27) /18(25) I/II/III/IV Vascular invasion, n(%) 29(54.7) /24(45.3) Yes/No
6
2. GEO database and TCGA_LIHC database analysis
To estimate the expression level of miR in HCC, public genomic data were obtained from The Cancer Genome Atlas liver hepatocellular carcinoma project (TCGA_LIHC, https://cancergenome.nih.gov) and the GEO database (GSE67140) (18) of the National Center for Biotechnology Information (NCBI).
3. Blood exosome Isolation and Total exosomal RNA Extraction
Human blood was collected from patients, centrifuged at 2,000 X g for 30 min at 4℃, at 10,000 X g for 45 min at 4℃. Exosomes were isolated from human blood samples using the ExoQuick (System Biosciences, Mountain View, CA, USA) according to the manufacturer’s protocol. RNA from blood-derived exosomes was extracted using the SeraMir Exosome RNA Amplification kit (System Biosciences, Palo Alto, USA). After isolation, total RNA concentration and purity were assessed using a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, USA).
4. Small RNA sequencing
Total RNA was isolated from exosomes of human blood using the SeraMir Exosome RNA Amplification kit (System Bioscience, Palo Alto, CA, USA) according to the
7
manufacturer’s instructions. Then, samples were quantitated with the NanoDrop-2000 spectrophotometer (Thermo Fisher Scientific, Cinisello Balsamo, Italy). The libraries were constructed from total RNA using the Illumina HiSeq 2000 system (Illumina Inc, San Diego, CA, USA), and only small RNAs ranging from 17 to 35 nucleotides were used to library construction.
5. Transmission electron microscopy
Transmission electron microscopy (TEM) was used to confirm the presence and size of exosomes. Sample fixation was conducted with 2% glutaraldehyde and 4% paraformaldehyde for 2 h at room temperature and responded with 0.13% methylcellulose and 0.4% uranyl acetate. Exosomes were then observed using Hitachi H-7600 TEM (Schaumburg).
6. Nanoparticle tracking analysis
Nanoparticle tracking analysis (NTA) was performed to the size distribution and concentration of exosomes using the properties of both light scattering and Brownian motion. NanoSight NS300 instrument (Malvern Panalytical Ltd., Malvern UK) equipped with a 405 nm laser was recorded with a frame rate of 30 frames/s, and the particle movement was evaluated using NTA software (version 3.0, Malvern Panalytical).
8
7. Western Blot
Proteins from cell lysates were prepared with radio immunoprecipitation (RIPA) buffer containing Halt Protease Inhibitor Cocktail (Thermo Fisher Scientific, Waltham, USA). Total proteins were separated by SDS-PAGE and transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore, Burlington, USA). The membranes were blocked in 5% non-fat milk in Tris-buffer saline and 0.1% Tween-20) and immunoblotted using the following primary antibodies: mouse anti-CD63 (1:1000, Abcam, Cambridge, USA), rabbit anti-CD9 (1:2000, Abcam, Cambridge, USA), mouse anti-CD81 (1:250, Invitrogen, Carlsbad, USA) and mouse anti-Bip/Grp78 (1:1000, BD, NJ, USA). Chemiluminescence signals were detected with Clarity™ Western ECL Substrate and ChemiDoc (both from BioRad, Hercules, USA).
8. Quantitative Real-time Polymerase chain reaction. (RT-qPCR)
The expression level of the blood-derived exo-miR was measured using RT-qPCR. The sequences of each miR were obtained in miRBase database (). Primer sequences used in the study are listed in supplementary Table 1. cDNA from exosome-derived RNA synthesis was performed using the miScript RT II kit (Qiagen, Hilden, Germany). RT-qPCR was performed using Amfisure qGreen qPCR Master Mix (Gendepot, Barker, TX, USA) and monitored in real time using CFX Connect™ Real-Time PCR Detection System (Bio-rad
9
Laboratories, Hercules, CA, USA). PCR conditions were following: 2min at 95℃, 40 cycles of 15s at 95℃, 34s at 58℃ or 60℃, and 30s at 72℃. Each sample was determined from triplicate reactions and normalized with has-miR-1228-3p. The relative standard curve
method (2-ΔΔCT) was used to determine relative expression. All measurements were
10 Table 2. Primer sequences used in this study.
Gene Accession No. Mature sequence Primer sequence
hsa-miR-106b MIMAT0000680 5’-UAAAGUGCUGACAGUGCAGAU –
3’
5'-TAAAGTGCTGACAGTGCAGAT –3'
hsa-miR-1307-5p MIMAT0022727 5’-UCGACCGGACCUCGACCGGCU-3’
5'-TCGACCGGACCTCGACCGGCT-3'
hsa-miR-340-5p MIMAT0004692
5’-UUAUAAAGCAAUGAGACUGAUU-3’
5'-TTATAAAGCAATGAGACTGATT-3'
11
9. Target prediction of miRNA
An in silico analysis was conducted to predict miR-1307-5p targeting genes by using the TargetScan 7.2 (http://www.targetscan.org/vert_72/).
10. Ingenuity pathway analysis (IPA)
Downstream pathway of miR-1307-5p and its’ target gene was analyzed with
functional annotation of EMT using Ingenuity pathway analysis (IPA) (QIAGEN Inc., United
States).
11. Statistical analysis
All experiments were performed at least three times and all samples were analyzed in triplicates. Statistical difference between each group was assessed by paired t-test or unpaired Welch’s t-test of Graphpad prism version 5.0 software (GraphPad Software lnc, San Diego, CA, USA). All differences were considered statistically significant at the level of P < 0.05. Overall survival (OS) and disease-free survival (DFS) were plotted using the Kaplan-Meier method and the significant differences were analyzed by the log-rank test. Statistical significance was established at P< 0.05.
12
III. RESULTS
1. Confirmation of isolated circulating exosomes and identification of overexpressed exo-miRs in metastasis group
Figure 1 demonstrates the pipeline of systematic integrative analyses performed in the present study for identifying circulating exo-miRs of metastatic driver potential. First, circulating exosomes were isolated and baseline characteristics of exosomes were evaluated. TEM revealed that the samples were consisted of 30–100 nm sized spherical vesicles (Figure 2A). The concentration and size distribution of the vesicles were also evaluated by NTA (Figure 2B). NTA verified that isolated particles were strongly enriched in the range of 30-100 nm. In western blot analyses, the isolated vesicles were positive for exosome markers (CD63, CD81, and CD9), and negative for Grp78, which considered as endoplasmic reticulum marker (Figure 2C). Base on the results, it was determined that the circulating exosomes were well isolated, and exosomal RNA was properly extracted. Small RNA sequencing libraries were successfully made in 14 samples among the 52 exosomal RNA preparation samples from the Plasma-HCC cohort. Fourteen samples were consisted of 8 from the metastasis-free group and 6 from the metastasis group. Sequencing data of plasma exo-miRs were analyzed and 61 predominantly overexpressed exo-miRs in the metastasis group were identified (> 2-fold, P < 0.05). Heatmap demonstrates overexpression of the 61 exo-miRs in the metastasis group. (Figure 2D)
15
2. Integrative analyses with public gene expression datasets to select potential candidates of pro-metastatic miR
For further selection of pro-metastatic miRs, systematic integrative analyses were performed with two different public gene expression datasets including GSE67140 and TCGA_LIHC. (18) The GSE67140 cohort is consisted of 91 HCCs with vascular invasion and 81 without vascular invasion. In the GSE67140 dataset, pattern analysis according to vascular invasion status was performed using CLICK algorithm. (19)
As a result, miRs could be categorized into 3 clusters by the expression pattern according to vascular invasion status (Figure 3A). Figure 3B illustrates heatmaps of the miR expression in each cluster according to vascular invasion status. Among the 3 clusters, 185 miRs of cluster 1 and 20 miRs of cluster 3 showed significantly higher expression value in vascular invasion group compared to that of non-vascular invasion group. Thus, the 205 miRs in cluster 1 or 3 were entered into the Venn diagram analysis. Venn diagram analysis revealed that nine miRs including miR-106b-5p, miR-1307-5p, miR-193b-3p, miR202-3p, miR33b-5p, miR-340-5p, miR-455-3p, miR-542-3p, and miR-574-3p were commonly overexpressed in both of the metastasis group and vascular invasion group in GSE67140 (Figure 3C).
The expression of the nine miRs in the 14-plasma exosome small RNA-seq dataset, were visualized according to metastasis status in Figure 3D. All of the nine miRs were significantly overexpressed in the metastasis group. Figure 3E demonstrates expression
16
of the candidate miRs according to vascular invasion status in GSE67140 cohort. All of the nine miRs were significantly overexpressed in vascular invasion group. Using TCGA_LIHC datasets, overall survival was analyzed according to expression of the nine miRs. (Figure 3E and Figure 4) Overexpression of 106b-5p, 1307-5p, or miR-340-5p were significantly associated with poor overall survival, while others did not show survival implication. Consequently, miR-106b-5p, miR-1307-5p, and miR-340-5p were selected as the potential candidates of metastatic driver in HCC.
20
3. Validating clinical implication of the candidate exo-miRs in the validation cohort
Expression of plasma exo-miR-106b-5p, miR-1307-5p, and miR-340-5p was measured in the entire Plasma-HCC cohort (N=52) to validate clinical implication of the selected miRs as a metastasis predictor. Figure 5A demonstrates expression of the three plasma exo-miRs in metastasis-free group (N=27) and metastasis group (N=25). Metastasis group showed significantly higher expression of plasma exo-miR-1307-5p compared to non-metastasis group (P=0.04), while expression of other two exo-miRs were not significantly different between two groups.
Further validation study to investigate clinical implication of the selected circulating exo-miRs was performed using Serum-HCC cohort due to the limited number and lack of clinical information of Plasma-HCC cohorts. Figure 5B shows expression of exo-miR-106b-5p, miR-1307-5p, and miR-340-5p according to vascular invasion status in the Serum-HCC cohort. The patients with vascular invasion showed significantly higher expression of serum exo-miR-106b-5p and miR-1307-5p. Figure 5C demonstrates the expression of serum miRs according to tumor recurrence status. The serum exo-miR-1307-5p was significantly overexpressed in tumor recurrence group. Figure 5D illustrates the expression of the three exo-miRs according to mUICC stages. The expression of serum exo-miR-1307-5p were gradually increased with progression of tumor stage (P< 0.0001), while expression of other two miRs did not show tumor stage-dependent relationship.
23
4. Predicting target genes of miR-1307-5p in HCC patients using bioinformatics analysis
We tried to identify downstream target genes of miR-1307-5p which promote extrahepatic metastasis. Target gene prediction using TargetScan 7.2 identified 120 candidates as miR-1307-5p target genes. (Figure 6A) Next, the expression of 120 genes were evaluated in TCGA_LIHC cohort. Because miRs negatively control their target genes, we tried to find significantly down regulated genes in HCC tissue. In TCGA_LIHC cohort, 16 of the 120 genes were down-regulated in HCC tissue compared to adjacent
non-tumor tissue, and 9 of 16 genes, including ALDH8A1, C11orf96, CLYBL, EFNB3, ENG,
NPC1L1, PIM3, SEC14L2, and SLC8A1, demonstrated statistical significance (P < 0.05). (Figure 6A and Figure 7)
To verify inversely correlated genes with miR 1307-5p, Pearson’s correlation analysis was performed using the expression data in TCGA_LIHC database. Expression of 5 of 9 genes, including ALDH8A1, C11orf96, CLYBL, ENG, and SEC14L2, showed inverse correlation with miR-1307-5p expression with statistical significance (r=<-0.3 and P < 0.05). (Figure 6B) We performed a pathway analysis with a functional annotation of the EMT using the IPA software on miR-1307-5p and the 5 target candidate gene (Figure 6C and Figure 8). As a consequence, it was confirmed that miR-1307-5p/SEC14L2/AKT and miR-1307-5p/ENG signal pathways were linked to EMT promotion. Survival analyses according to expression of ENG and SEC14L2 were performed using expression data in
24
TCGA_LIHC. Figure 6D and Figure 6E illustrates Kaplan-Meier plot of OS and DFS according to expression of ENG and SEC14L2, respectively. Low ENG expression group showed significantly poor OS (P=0.0002) and poor DFS survival (P=0.0026) compared to high expression group. In addition to ENG, low SEC14L2 group demonstrated poor OS (P=0.011) and DFS (P=0.003) compared to high expression group.
29
Ⅳ. DISSCUSSION
A growing body of evidence indicates that exosomes transfer pro-metastatic molecules to recipient cells, thereby resulting pre-metastatic niche formation. (14, 15) The present study was performed under the assumption that the expression of specific exo-miRs would be increased in systemic circulation prior to extrahepatic metastasis and they could play as a driver of metastasis in patients with HCCs.
To confirm the assumption, we analyzed differentially expressed circulating exo-miR profiles between metastasis-free group and metastasis group during follow-up. Among the 61 predominantly over-expressed plasma exo-miRs in the metastasis group, further selection of candidate miRs were performed by systematic integrative analyses with publicly available RNA-seq datasets. Consequently, miR-106b-5p, miR-1307-5p and miR-340-5p, were selected as potential candidates for pro-metastatic miRs. In validation study, metastasis group demonstrates significantly overexpressed circulating exo-miR-1307-5p. Furthermore, down regulation of SEC14L2 and ENG and promoting EMT were proposed as possible downstream pathway of miR-1307-5p in comprehensive bioinformatics analyses. In our knowledge, this is the first study which identified circulating exo-miR-1307-5p as a novel metastasis driver molecule and metastasis predictor in HCC patients.
Previous studies reported that exosomes enclose unique cargoes from parent cell and the exosomal cargoes have been highlighted as a promising biomarker in cancer
30
biology. Recently, several studies reported circulating exo-miRs as potential diagnostic biomarker for early stage HCC. (20, 21) Moreover, studies have shown that the expression of an abnormally regulated exo-miRs can promote HCC progression and metastasis, by altering the genetic network. (22) Lin et al. reported that exosome-mediated miR delivery promotes HCC EMT and metastasis in cell line study. (23) However, few studies have sought to identify circulating exo-miR profile as metastasis predictor and metastasis driver in HCC. In the present study, circulating exo-miR-1307-5p was identified as a potential candidate of metastasis predictor and metastasis driving molecule in HCC patients. Previously, miR-1307 has been reported as onco-miR in diverse cancers and
serum exo-miR-1307 has been reported as ovarian cancer biomarker. (24-26) In HCC,
one recent study reported that miR-1307 promote HCC growth and metastasis by inhibiting DAB2 interacting protein in HCC. (27) Although the present study identified exo-miR-1307-5p as a potential candidate of metastasis driver, further mechanism study is necessary to identify how circulating exo-miR-1307-5p promotes HCC metastasis and whether it could be potential therapeutic target of HCC.
Presence of vascular invasion has been considered as an important hallmark of HCC invasiveness and poor prognosis. (28) It also has been reported as a principal predictive marker for extra-hepatic metastasis of HCC. (29) In the current study, all of the nine candidate miRs were significantly overexpressed in patients with metastasis as well as vascular invasion. Considering vascular invasion is closely associated with subsequent extrahepatic metastasis in patients with HCC, exo-miR-1307-5p might be
31
predictive marker for extrahepatic metastasis and/or vascular invasion.
EMT is a process that loses the characteristics of epithelial cells and acquires the mesenchymal phenotype. (30) EMT plays a crucial role in promoting tumor invasiveness and metastasis by inducing the loss of cell to cell adhesion. (31) In the present study, we proposed SEC14L2/Akt and ENG-related signaling pathways as downstream pathway of miR-1307-5p for promoting EMT in HCC patients. According to pathway analysis using IPA, SEC14L2 is down-regulated by miR-1307, which in turn leads to Akt pathway activation, promoting EMT. SEC14L2 has been reported as a potent tumor suppressor gene in various malignancies. (32) In HCC, Li et al reported that SEC14L2, as a novel master regulator gene, exerted an anti-proliferative effect in HCC cells and strongly suppressed tumor growth in a mouse model. (33) ENG (CD105), a transmembrane glycoprotein, is one of the transforming growth factor β co-receptors. (34) ENG is involved in angiogenesis of solid tumors including HCC. (35) Several studies reported that down-regulation of ENG in HCC tissue, as well as serum concentrations, could play as a poor prognostic marker for HCC patients. (34, 36, 37) However, the molecular mechanisms of ENG in HCC progression are still poorly understood. According to the IPA analysis performed in this study, down regulation of ENG by miR-1307-5p promotes EMT by activating PLAU, RHOA, SMAD3, TWIST1 and MYC. Further mechanism research is needed to verify the downstream pathways proposed by the IPA.
This study has two limitations. The first limitation is the small number of the included patients. In our study, metastasis group was defined as patients who have
32
developed extrahepatic metastasis after initial blood sampling point. Because HCC patients with the occurrence of metastasis who had available blood samples were rare, we could not enroll sufficient number of patients for powerful statistical analysis. From different points of view, due to the shortage of blood sample of metastatic HCC patients, we assume that the results of current study provide valuable and potentially useful information about pro-metastatic exo-miR. Further validation study in larger cohort is needed to verify the result of this study. Second, mechanism study of how exo-miR-1307-5p could facilitate extra-hepatic metastasis could not be performed. To overcome
this limitation, we implemented in silico analytic strategy. MiR-1307-5p downstream
pathway promoting EMT was predicted by using bioinformatics analyses. Experimental validation to verify the involved signaling pathway would be required as well.
In conclusion, our present study revealed circulating exo-miR-1307-5p as a predictive marker for the occurrence of metastasis in HCC patients using systematic integrative analyses. EMT promotion by down regulation of SEC14L2 and ENG could be possible downstream pathway of miR-1307-5p by comprehensive bioinformatics analyses.
33
V. REFERENCE
1. Ghouri YA, Mian I, Rowe JH. Review of hepatocellular carcinoma: Epidemiology,
etiology, and carcinogenesis. J Carcinog. 2017;16:1.
2. Wong R, Frenette C. Updates in the management of hepatocellular carcinoma.
Gastroenterol Hepatol (N Y). 2011;7(1):16-24.
3. Crissien AM, Frenette C. Current management of hepatocellular carcinoma.
Gastroenterol Hepatol (N Y). 2014;10(3):153-61.
4. Heimbach JK, Kulik LM, Finn RS, Sirlin CB, Abecassis MM, Roberts LR, et al. AASLD
guidelines for the treatment of hepatocellular carcinoma. Hepatology. 2018;67(1):358-80.
5. Cheung AH, Chow C, To KF. Latest development of liquid biopsy. J Thorac Dis.
2018;10(Suppl 14):S1645-S51.
6. Diaz LA, Jr., Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin
Oncol. 2014;32(6):579-86.
7. Buder A, Tomuta C, Filipits M. The potential of liquid biopsies. Curr Opin Oncol.
2016;28(2):130-4.
8. Gold B, Cankovic M, Furtado LV, Meier F, Gocke CD. Do circulating tumor cells,
exosomes, and circulating tumor nucleic acids have clinical utility? A report of the association for molecular pathology. J Mol Diagn. 2015;17(3):209-24.
34 investigation. Methods. 2015;87:31-45.
10. Bebelman MP, Smit MJ, Pegtel DM, Baglio SR. Biogenesis and function of
extracellular vesicles in cancer. Pharmacol Ther. 2018;188:1-11.
11. Contreras-Naranjo JC, Wu HJ, Ugaz VM. Microfluidics for exosome isolation and
analysis: enabling liquid biopsy for personalized medicine. Lab Chip. 2017;17(21):3558-77.
12. Zhang J, Li S, Li L, Li M, Guo C, Yao J, et al. Exosome and exosomal microRNA:
trafficking, sorting, and function. Genomics Proteomics Bioinformatics. 2015;13(1):17-24.
13. Safdar A, Saleem A, Tarnopolsky MA. The potential of endurance exercise-derived
exosomes to treat metabolic diseases. Nat Rev Endocrinol. 2016;12(9):504-17.
14. Syn N, Wang L, Sethi G, Thiery JP, Goh BC. Exosome-Mediated Metastasis: From
Epithelial-Mesenchymal Transition to Escape from Immunosurveillance. Trends Pharmacol Sci. 2016;37(7):606-17.
15. Greening DW, Gopal SK, Mathias RA, Liu L, Sheng J, Zhu HJ, et al. Emerging roles
of exosomes during epithelial-mesenchymal transition and cancer progression. Semin Cell Dev Biol. 2015;40:60-71.
16. Kim TH, Kim SY, Tang A, Lee JM. Comparison of international guidelines for
noninvasive diagnosis of hepatocellular carcinoma: 2018 update. Clin Mol Hepatol. 2019;25(3):245-63.
17. Korean Liver Cancer A, National Cancer Center GK. 2018 Korean Liver Cancer
35
Hepatocellular Carcinoma. Korean J Radiol. 2019;20(7):1042-113.
18. Xie QY, Almudevar A, Whitney-Miller CL, Barry CT, McCall MN. A microRNA
biomarker of hepatocellular carcinoma recurrence following liver transplantation accounting for within-patient heterogeneity. BMC Med Genomics. 2016;9:18.
19. Sharan R, Maron-Katz A, Shamir R. CLICK and EXPANDER: a system for clustering
and visualizing gene expression data. Bioinformatics. 2003;19(14):1787-99.
20. Cho HJ, Eun JW, Baek GO, Seo CW, Ahn HR, Kim SS, et al. Serum Exosomal
MicroRNA, miR-10b-5p, as a Potential Diagnostic Biomarker for Early-Stage Hepatocellular Carcinoma. J Clin Med. 2020;9(1).
21. Cho HJ, Baek GO, Seo CW, Ahn HR, Sung S, Son JA, et al. Exosomal
microRNA-4661-5p-based serum panel as a potential diagnostic biomarker for early-stage hepatocellular carcinoma. Cancer Med. 2020.
22. Qu Z, Wu J, Wu J, Ji A, Qiang G, Jiang Y, et al. Exosomal miR-665 as a novel
minimally invasive biomarker for hepatocellular carcinoma diagnosis and prognosis. Oncotarget. 2017;8(46):80666-78.
23. Lin Q, Zhou CR, Bai MJ, Zhu D, Chen JW, Wang HF, et al. Exosome-mediated
miRNA delivery promotes liver cancer EMT and metastasis. Am J Transl Res. 2020;12(3):1080-95.
24. Tang R, Qi Q, Wu R, Zhou X, Wu D, Zhou H, et al. The polymorphic terminal-loop
of pre-miR-1307 binding with MBNL1 contributes to colorectal carcinogenesis via interference with Dicer1 recruitment. Carcinogenesis. 2015;36(8):867-75.
36
25. Han S, Zou H, Lee JW, Han J, Kim HC, Cheol JJ, et al. miR-1307-3p Stimulates
Breast Cancer Development and Progression by Targeting SMYD4. J Cancer. 2019;10(2):441-8.
26. Su YY, Sun L, Guo ZR, Li JC, Bai TT, Cai XX, et al. Upregulated expression of serum
exosomal miR-375 and miR-1307 enhance the diagnostic power of CA125 for ovarian cancer. J Ovarian Res. 2019;12(1):6.
27. Chen S, Wang L, Yao B, Liu Q, Guo C. miR-1307-3p promotes tumor growth and
metastasis of hepatocellular carcinoma by repressing DAB2 interacting protein. Biomed Pharmacother. 2019;117:109055.
28. European Association For The Study Of The L, European Organisation For R,
Treatment Of C. EASL-EORTC clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol. 2012;56(4):908-43.
29. Hashimoto M, Kobayashi T, Ishiyama K, Ide K, Ohira M, Tahara H, et al. Predictive
Independent Factors for Extrahepatic Metastasis of Hepatocellular Carcinoma Following Curative Hepatectomy. Anticancer Res. 2017;37(5):2625-31.
30. Blackwell RH, Foreman KE, Gupta GN. The Role of Cancer-Derived Exosomes in
Tumorigenicity & Epithelial-to-Mesenchymal Transition. Cancers (Basel). 2017;9(8).
31. Gotzmann J, Mikula M, Eger A, Schulte-Hermann R, Foisner R, Beug H, et al.
Molecular aspects of epithelial cell plasticity: implications for local tumor invasion and metastasis. Mutat Res. 2004;566(1):9-20.
37
tocopherol-associated protein (TAP/Sec14L2) in human breast cancer. Cancer Invest. 2009;27(10):971-7.
33. Li Z, Lou Y, Tian G, Wu J, Lu A, Chen J, et al. Discovering master regulators in
hepatocellular carcinoma: one novel MR, SEC14L2 inhibits cancer cells. Aging (Albany NY). 2019;11(24):12375-411.
34. Kasprzak A, Adamek A. Role of Endoglin (CD105) in the Progression of
Hepatocellular Carcinoma and Anti-Angiogenic Therapy. Int J Mol Sci. 2018;19(12).
35. Ho JW, Poon RT, Sun CK, Xue WC, Fan ST. Clinicopathological and prognostic
implications of endoglin (CD105) expression in hepatocellular carcinoma and its adjacent non-tumorous liver. World J Gastroenterol. 2005;11(2):176-81.
36. Yagmur E, Rizk M, Stanzel S, Hellerbrand C, Lammert F, Trautwein C, et al. Elevation
of endoglin (CD105) concentrations in serum of patients with liver cirrhosis and carcinoma. Eur J Gastroenterol Hepatol. 2007;19(9):755-61.
37. Teama S, Fawzy A, Teama S, Helal A, Drwish AD, Elbaz T, et al. Increased Serum
Endoglin and Transforming Growth Factor beta1 mRNA Expression and Risk of Hepatocellular Carcinoma in Cirrhotic Egyptian Patients. Asian Pac J Cancer Prev. 2016;17(5):2429-34.
38 국문요약 혈액 내 엑소좀 miR-1307-5p는 간암 전이에 잠재적인 마커에 대한 연구 아주대학교 대학원 의생명과학과 서 철 원 (지도교수: 정 재 연) 간암은 전 세계적으로 5대 암으로 불릴 정도로 위험한 암으로 알려져 있다. 또 한 간암은 수술적 절제 후에도 재발 및 전이가 되기 쉽다. 현재 간암을 진단하는데 사 용되고 있는 알파태아단백(AFP)는 민감도나 정확도가 많이 떨어지고 있다. 또한 간암 전이에 관한 진단은 어려움을 겪고 있다. 이에 있어 간암 전이를 진단 및 예측하는데 있어 새로운 방법이 필요한 시점이다. 현재 microRNA는 바이오 마커로 많이 연구되고 있는 부분이며, 최근에는 엑소좀 안에 있는 microRNA로써 새로운 바이오 마커를 연구 하고 있다. 엑소좀은 30nm-150nm의 작은 소포체로써 RNA, microRNA 뿐만 아니라 지 질, 단백질, 핵산 등을 포함하고 세포 간 신호전달 물질로 각광받고 있다. 엑소좀 안에 있는 microRNA를 이용하여 간암 전이 마커를 확인해보고자 하였다.
39
본 연구에서는 정상군 환자와 간암환자의 혈청과 혈장을 이용하였고, 간암환자 중에서도 전이가 있었던 환자와 전이가 없었던 환자를 분류하였다. 환자의 혈청과 혈 장에서 엑소좀을 추출하여 전사체 분석을 진행 하였고, 그 외에 GEO data와 TCGA data를 이용하여 간암 전이에 연관이 있는 엑소좀 microRNA 후보를 확인 하였다. 선 별된 엑소좀 microRNA 후보는 각 환자별로 실시간 중합효소 연쇄반응 실험을 통해 전 이와 밀접한 관련이 있는 엑소좀 microRNA를 선별하였다. 최종 선별된 엑소좀 miR-1307-5에 대하여 연관 있다고 예상되는 유전자를 확인 하였고 더 나아가 관련 유전자 와의 신호 전달 경로까지 확인한 결과 엑소좀 miR-1307-5p가 ENG를 막으면 여러 신 호 전달 경로를 통해 최종적으로는 EMT를 촉진한다고 예측되었다. 결과적으로 우리의 연구는 엑소좀 microRNA를 이용하여 간암 전이 관련 잠재 적인 바이오 마커로써의 가능성을 확인 하였고, 더 나아가 간암 전이 진단에 새로운 가능성을 기대한다. 핵심어 : 간세포암, 전이, 엑소좀, microRNA, bioinformatics 분석